Abstract
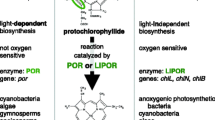
Although the plastid genomes of diatoms maintain a conserved architecture and core gene set, considerable variation about this core theme exists and can be traced to several different processes. Gene duplication, pseudogenization, and loss, as well as intracellular transfer of genes to the nuclear genome, have all contributed to variation in gene content among diatom species. In addition, some noncoding sequences have highly restricted phylogenetic distributions that suggest a recent foreign origin. We sequenced the plastid genome of the marine diatom, Toxarium undulatum, and found that the genome contains three genes (chlB, chlL, and chlN) involved in light-independent chlorophyll a biosynthesis that were not previously known from diatoms. Phylogenetic and syntenic data suggest that these genes were differentially retained in this one lineage as they were repeatedly lost from most other diatoms. Unique among diatoms and other heterokont algae sequenced so far, the genome also contains a large group II intron within an otherwise intact psaA gene. Although the intron is most similar to one in the plastid-encoded psaA gene of some green algae, high sequence divergence between the diatom and green algal introns rules out recent shared ancestry. We conclude that the psaA intron was likely introduced into the plastid genome of T. undulatum, or some earlier ancestor, by horizontal transfer from an unknown donor. This genome further highlights the myriad processes driving variation in gene and intron content in the plastid genomes of diatoms, one of the world’s foremost primary producers.



Similar content being viewed by others
References
Adachi J, Waddell JP, Martin W, Hasegawa M (2000) Plastid genome phylogeny and a model of amino acid substitution for proteins encoded by chloroplast DNA. J Mol Evol 50:348–358
Amin SA, Parker MS, Armbrust EV (2012) Interactions between diatoms and bacteria. Microbiol Mol Biol Rev 76:667–684
Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kroger N, Lau WWY, Lane TW, Larimer FW, Lippmeier JC, Lucas S, Medina M, Montsant A, Obornik M, Parker MS, Palenik B, Pazour GJ, Richardson PM, Rynearson TA, Saito MA, Schwartz DC, Thamatrakoln K, Valentin K, Vardi A, Wilkerson FP, Rokhsar DS (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306:79–86
Armstrong GA (1998) Greening in the dark: light-independent chlorophyll biosynthesis from anoxygenic photosynthetic bacteria to gymnosperms. J Photochem Photobiol B 43:87–100
Belfort M, Perlman PS (1995) Mechanisms of intron mobility. J Biol Chem 270:30237–30240
Bendich AJ (2004) Circular chloroplast chromosomes: the grand illusion. Plant Cell 16:1661–1666
Blocker FJ, Mohr G, Conlan LH, Qi LI, Belfort M, Lambowitz AM (2005) Domain structure and three-dimensional model of a group II intron-encoded reverse transcriptase. RNA 11:14–28
Boisvert S, Laviolette F, Corbeil J (2010) Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol 17:1519–1533
Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, Rayko E, Salamov A, Vandepoele K, Beszteri B, Gruber A, Heijde M, Katinka M, Mock T, Valentin K, Verret F, Berges JA, Brownlee C, Cadoret JP, Chiovitti A, Choi CJ, Coesel S, De Martino A, Detter JC, Durkin C, Falciatore A, Fournet J, Haruta M, Huysman MJ, Jenkins BD, Jiroutova K, Jorgensen RE, Joubert Y, Kaplan A, Kroger N, Kroth PG, La Roche J, Lindquist E, Lommer M, Martin-Jezequel V, Lopez PJ, Lucas S, Mangogna M, McGinnis K, Medlin LK, Montsant A, Oudot-Le Secq MP, Napoli C, Obornik M, Parker MS, Petit JL, Porcel BM, Poulsen N, Robison M, Rychlewski L, Rynearson TA, Schmutz J, Shapiro H, Siaut M, Stanley M, Sussman MR, Taylor AR, Vardi A, von Dassow P, Vyverman W, Willis A, Wyrwicz LS, Rokhsar DS, Weissenbach J, Armbrust EV, Green BR, Van de Peer Y, Grigoriev IV (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456:239–244
Boyd PW, Jickells T, Law CS, Blain S, Boyle EA, Buesseler KO, Coale KH, Cullen JJ, de Baar HJ, Follows M, Harvey M, Lancelot C, Levasseur M, Owens NP, Pollard R, Rivkin RB, Sarmiento J, Schoemann V, Smetacek V, Takeda S, Tsuda A, Turner S, Watson AJ (2007) Mesoscale iron enrichment experiments 1993-2005: synthesis and future directions. Science 315:612–617
Brembu T, Winge P, Tooming-Klunderud A, Nederbragt AJ, Jakobsen KS, Bones AM (2014) The chloroplast genome of the diatom Seminavis robusta: new features introduced through multiple mechanisms of horizontal gene transfer. Mar Genom 16:17–27
Deschamps P, Moreira D (2012) Reevaluating the green contribution to diatom genomes. Genome Biol Evol 4:683–688
Fong A, Archibald JM (2008) Evolutionary dynamics of light-independent protochlorophyllide oxidoreductase genes in the secondary plastids of cryptophyte algae. Eukaryot Cell 7:550–553
Gordon PM, Piccirilli JA (2001) Metal ion coordination by the AGC triad in domain 5 contributes to group II intron catalysis. Nat Struct Biol 8:893–898
Guillard RRL (1975) Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH (eds) Culture of marine invertebrate animals. Plenum Press, New York, pp 26–60
Hasle GR, Syvertsen EE (1997) Marine diatoms. In: Tomas CR (ed) Identifying marine phytoplankton. Academic Press, San Diego, pp 5–386
Hunsperger HM, Randhawa T, Cattolico RA (2015) Extensive horizontal gene transfer, duplication, and loss of chlorophyll synthesis genes in the algae. BMC Evol Biol 15:1–19
Johansen SD, Haugen P, Nielsen H (2007) Expression of protein-coding genes embedded in ribosomal DNA. Biol Chem 388:679–686
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Khan H, Archibald JM (2008) Lateral transfer of introns in the cryptophyte plastid genome. Nucleic Acids Res 36:3043–3053
Kooistra WHCF, De Stefano M, Mann DG, Salma N, Medlin LK (2003) Phylogenetic position of Toxarium, a pennate-like lineage within centric diatoms (Bacillariophyceae). J Phycol 39:185–197
Lambowitz AM, Belfort M (1993) Introns as mobile genetic elements. Annu Rev Biochem 62:587–622
Laslett D, Canback B (2004) ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16
Lemieux C, Otis C, Turmel M (2014) Chloroplast phylogenomic analysis resolves deep-level relationships within the green algal class Trebouxiophyceae. BMC Evol Biol 14:1–15
Lemieux C, Vincent AT, Labarre A, Otis C, Turmel M (2015) Chloroplast phylogenomic analysis of chlorophyte green algae identifies a novel lineage sister to the Sphaeropleales (Chlorophyceae). BMC Evol Biol 15:1–13
Lohse M, Drechsel O, Bock R (2007) OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet 52:267–274
Lommer M, Roy AS, Schilhabel M, Schreiber S, Rosenstiel P, LaRoche J (2010) Recent transfer of an iron-regulated gene from the plastid to the nuclear genome in an oceanic diatom adapted to chronic iron limitation. BMC Genom 11:718. doi:10.1186/1471-2164-11-718
Lommer M, Specht M, Roy A-S, Kraemer L, Andreson R, Gutowska M, Wolf J, Bergner S, Schilhabel M, Klostermeier U, Beiko R, Rosenstiel P, Hippler M, LaRoche J (2012) Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biol 13:R66
Mann DG, Vanormelingen P (2013) An inordinate fondness? The number, distributions, and origins of diatom species. J Eukaryot Microbiol 60:414–420
Marchetti A, Parker MS, Moccia LP, Lin EO, Arrieta AL, Ribalet F, Murphy MEP, Maldonado MT, Armbrust EV (2009) Ferritin is used for iron storage in bloom-forming marine pennate diatoms. Nature 457:467–470
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:D222–D226
Medlin LK, Sato S, Mann DG, Kooistra WHCF (2008) Molecular evidence confirms sister relationship of Ardissonea, Climacosphenia, and Toxarium within the bipolar centric diatoms (Bacillariophyta, Mediophyceae), and cladistic analyses confirm that extremely elongated shape has arisen twice in the diatoms. J Phycol 44:1340–1348
Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D (2009) Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324:1724–1726
Mower J, Stefanovic S, Hao W, Gummow J, Jain K, Ahmed D, Palmer J (2010) Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biol 8:150
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274
Peers G, Price NM (2006) Copper-containing plastocyanin used for electron transport by an oceanic diatom. Nature 441:341–344
Perrineau M-M, Price DC, Mohr G, Bhattacharya D (2015) Recent mobility of plastid encoded group II introns and twintrons in five strains of the unicellular red alga Porphyridium. PeerJ 3:e1017
Qu G, Kaushal PS, Wang J, Shigematsu H, Piazza CL, Agrawal RK, Belfort M, Wang H-W (2016) Structure of a group II intron in complex with its reverse transcriptase. Nat Struct Mol Biol 23:549–557
Raymond J, Siefert JL, Staples CR, Blankenship RE (2004) The natural history of nitrogen fixation. Mol Biol Evol 21:541–554
Reinbothe S, Reinbothe C, Apel K, Lebedev N (1996) Evolution of chlorophyll biosynthesis—the challenge to survive photooxidation. Cell 86:703–705
Rice DW, Alverson AJ, Richardson AO, Young GJ, Sanchez-Puerta MV, Munzinger J, Barry K, Boore JL, Zhang Y, Knox EB (2013) Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 342:1468–1473
Round FE, Crawford RM, Mann DG (1990) The diatoms: biology and morphology of the genera. Cambridge University Press, Cambridge
Ruck EC, Nakov T, Jansen RK, Theriot EC, Alverson AJ (2014) Serial gene losses and foreign DNA underlie size and sequence variation in the plastid genomes of diatoms. Genome Biol Evol 6:644–654
Sabir JSM, Yu M, Ashworth MP, Baeshen NA, Baeshen MN, Bahieldin A, Theriot EC, Jansen RK (2014) Conserved gene order and expanded inverted repeats characterize plastid genomes of Thalassiosirales. PLoS One 9:e107854
Salzberg SL, White O, Peterson J, Eisen JA (2001) Microbial genes in the human genome: lateral transfer or gene loss? Science 292:1903–1906
Sánchez-Puerta MV, Bachvaroff TR, Delwiche CF (2005) The complete plastid genome sequence of the haptophyte Emiliania huxleyi: a comparison to other plastid genomes. DNA Res 12:151–156
Sánchez-Puerta MV, Cho Y, Mower JP, Alverson AJ, Palmer JD (2008) Frequent, phylogenetically local horizontal transfer of the cox1 group I Intron in flowering plant mitochondria. Mol Biol Evol 25:1762–1777
Simola DF, Wissler L, Donahue G, Waterhouse RM, Helmkampf M, Roux J, Nygaard S, Glastad KM, Hagen DE, Viljakainen L, Reese JT, Hunt BG, Graur D, Elhaik E, Kriventseva EV, Wen J, Parker BJ, Cash E, Privman E, Childers CP, Muñoz-Torres MC, Boomsma JJ, Bornberg-Bauer E, Currie CR, Elsik CG, Suen G, Goodisman MAD, Keller L, Liebig J, Rawls A, Reinberg D, Smith CD, Smith CR, Tsutsui N, Wurm Y, Zdobnov EM, Berger SL, Gadau J (2013) Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. Genome Res 23:1235–1247
Sloan DB, Nakabachi A, Richards S, Qu J, Murali SC, Gibbs RA, Moran NA (2014) Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol 31:857–871
Smith DR (2012) Updating our view of organelle genome nucleotide landscape. Front Genet 3:175
Smith DR, Keeling PJ (2015) Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. P Natl Acad Sci USA 112:10177–10184
Sorhannus U (2007) A nuclear-encoded small-subunit ribosomal RNA timescale for diatom evolution. Mar Micropaleontol 65:1–12
Strzepek RF, Harrison PJ (2004) Photosynthetic architecture differs in coastal and oceanic diatoms. Nature 431:689–692
Tajima N, Saitoh K, Sato S, Maruyama F, Ichinomiya M, Yoshikawa S, Kurokawa K, Ohta H, Tabata S, Kuwata A, Sato N (2016) Sequencing and analysis of the complete organellar genomes of Parmales, a closely related group to Bacillariophyta (diatoms). Curr Genet. doi:10.1007/s00294-016-0598-y
Theriot EC, Ashworth MP, Nakov T, Ruck E, Jansen RK (2015) Dissecting signal and noise in diatom chloroplast protein encoding genes with phylogenetic information profiling. Mol Phylogenet Evol 89:28–36
Tomaru Y, Nagasaki K (2011) Diatom viruses. In: Seckbach J, Kociolek P (eds) The diatom world. Springer, Netherlands, pp 211–225
Turmel M, Cote V, Otis C, Mercier JP, Gray MW, Lonergan KM, Lemieux C (1995) Evolutionary transfer of ORF-containing group I introns between different subcellular compartments (chloroplast and mitochondrion). Mol Biol Evol 12:533–545
Ueda M, Tanaka A, Sugimoto K, Shikanai T, Nishimura Y (2014) chlB requirement for chlorophyll biosynthesis under short photoperiod in Marchantia polymorpha L. Genome Biol Evol 6:620–628
Woehle C, Dagan T, Martin WF, Gould SB (2011) Red and problematic green phylogenetic signals among thousands of nuclear genes from the photosynthetic and apicomplexa-related Chromera velia. Genome Biol Evol 3:1220–1230
Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255
Yamazaki S, Nomata J, Fujita Y (2006) Differential operation of dual protochlorophyllide reductases for chlorophyll biosynthesis in response to environmental oxygen levels in the cyanobacterium Leptolyngbya boryana. Plant Physiol 142:911–922
Yang Z (1995) A space-time process model for the evolution of DNA sequences. Genetics 139:993–1005
Zimmerly S, Semper C (2015) Evolution of group II introns. Mob DNA 6:1–19
Acknowledgments
We thank Colton Kessenich for help with the genome assembly; David Chafin, Jeff Pummill, and Pawel Wolinski for providing computational support through the Arkansas High Performance Computing Center (AHPCC); and Suresh Kumar and Jeffrey Lewis for critical comments on an earlier version of the manuscript. This material is based upon work supported by the National Science Foundation (NSF) under Grant No. DEB-1353131 and an award from the Arkansas Biosciences Institute. This research used computational resources available through the AHPCC, which is funded through multiple NSF grants and the Arkansas Economic Development Commission.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Electronic supplementary material
Below is the link to the electronic supplementary material.
294_2016_652_MOESM1_ESM.pdf
Figure S1. Individual gene phylogenies based on amino acid alignments for the chlB, chlL, and chlN genes. Numbers at nodes are standard bootstrap proportions from 500 pseudoreplicates (PDF 22 kb)
Rights and permissions
About this article
Cite this article
Ruck, E.C., Linard, S.R., Nakov, T. et al. Hoarding and horizontal transfer led to an expanded gene and intron repertoire in the plastid genome of the diatom, Toxarium undulatum (Bacillariophyta). Curr Genet 63, 499–507 (2017). https://doi.org/10.1007/s00294-016-0652-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-016-0652-9