Abstract
Cell fate decisions are controlled by multiple cell-intrinsic and -extrinsic factors. In budding yeast, the decision to enter gametogenesis or sporulation is dictated by nutrient availability and mating type. Recently, we showed that in diploid cells harbouring opposite mating types (MATa and MATα), the protein kinase A (PKA) and target of rapamycin complex I (TORC1) signalling pathways integrate at the promoter of the master regulatory transcription factor IME1 to control sporulation via nutrient availability (Weidberg, et al. 2016). In cells with a single mating type (MATa or MATα), however, IME1 is repressed by transcription through the IME1 promoter of a long non-coding RNA called IRT1, which prevents this cell type from undergoing sporulation. Here, we investigated the role of nutrient signalling in mating-type control of IME1. We find that expression of IRT1, like IME1 itself, depends on nutrient availability and the activities of PKA and TORC1. IRT1 transcription is repressed when nutrients are ample and TORC1 and PKA are active. In contrast, inhibition of PKA and TORC1 is sufficient to recruit Rme1 to the IRT1 promoter and induce IRT1-mediated repression of IME1. Finally, we provide evidence that IRT1 and IME1 are co-repressed by the Tup1–Cyc8 complex when nutrients are ample. Thus, in cells with a single mating-type nutrient availability regulates mating-type repression of IME1 and sporulation. Our results indicate that there is a hierarchy between nutrient and mating-type signals in controlling the decision to enter sporulation.
Similar content being viewed by others
Introduction
Cell fate decisions are regulated by a multitude of intrinsic and extrinsic signals. How multiple signals are integrated to make a binary cell fate choice is often not well understood.
Budding yeast gametogenesis or sporulation is an ideal model system to study this problem. When the signal requirements to enter sporulation are met, cells induce a gene expression cascade to generate four haploid spores from a diploid cell (Honigberg and Purnapatre 2003; Jin and Neiman 2016; Kassir et al. 2003; van Werven and Amon 2011; Vershon and Pierce 2000).
Nutrient and mating-type signals control the decision leading to meiosis and gamete or spore formation. These signals integrate at the promoter of the master regulator for entry into sporulation, called IME1 (Kassir et al. 1988; Nachman et al. 2007). When nutrients are ample, IME1 is repressed because the two major nutrient sensing signalling pathways, protein kinase A and target of rapamycin complex I (TORC1), are active (Weidberg et al. 2016). In the absence of nitrogen and glucose compounds in the growth medium, TORC1 and PKA activities are repressed and IME1 is strongly induced.
The mating-type loci also control entry into sporulation. IME1 can only be induced in diploid cells because the a1–α2 complex, which expression requires both mating loci (MATa and MATα), represses the transcription of RME1 (Mitchell and Herskowitz 1986). Cells with a single mating type (MATa or MATα), however, exhibit high levels of Rme1 which in turn represses IME1 via an unusual mechanism (Covitz and Mitchell 1993; van Werven et al. 2012). Rme1 activates the transcription of the long non-coding RNA IRT1, which is expressed from upstream in the IME1 promoter and spans almost the complete promoter. Repression of IME1 by IRT1 transcription requires repressive chromatin marks such as histone H3 lysine 36 methylation and lysine 4 di-methylation (van Werven et al. 2012).
How nutrient availability regulates IRT1 transcription is not well understood. Here, we demonstrate that IRT1 transcription, like IME1, is under control of the TORC1 and PKA nutrient sensing pathways. In addition, we provide evidence that the transcriptional repressor Tup1–Cyc8, like IME1, represses IRT1. We propose that co-repression of IME1 and IRT1 serves as a fail-safe mechanism for mating-type control of sporulation. Overall, our results suggest that a hierarchy between nutrient and mating-type signals controls the decision to enter sporulation.
Materials and methods
Strains
SK1 strain background was used for the experiments throughout this manuscript. The tup1Δ strain was generated using one-step deletion protocol as described by Longtine et al. (1998). The tpk1as and Rme1-V5 alleles were described previously (van Werven et al. 2012; Weidberg et al. 2016). The genotypes of the strains used for this study are described in Table 1.
Growth conditions
For Fig. 1, cells were grown overnight in YPD (1 % yeast extract, 2 % peptone, 2 % glucose) at 30 °C, then diluted to fresh YPD (OD600 = 1) and treated with different drugs. For Fig. 2, cells were grown overnight and diluted to OD600 = 2, and samples were taken at the indicated time points.
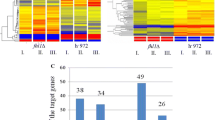
IRT1 is induced in rich medium when PKA and TORC1 are inhibited. a Overview of IRT1/IME1 locus (top), and northern blot of IRT1 expression (bottom). Haploid cells harbouring the tpk1M164G, tpk2Δ, tpk3Δ alleles (tpk1as) (FW1760) were grown overnight in YPD, diluted into YPD plus 1NM-PP1 or rapamycin/1NM-PP1, and samples were taken at the indicated time points. Total RNA was isolated, separated by gel electrophoresis, blotted and probed for IRT1. b IRT1 RNA quantification in haploid cells harbouring tpk1as (FW1760). Cells were grown overnight in YPD, diluted into fresh YPD in absence or presence of rapamycin, 1NM-PP1, or rapamycin/1NM-PP1. Samples were taken at the indicated time points. Total RNA was isolated, reverse transcribed, and IRT1 mRNA levels were measured by quantitative PCR. Signals were normalized to ACT1 levels. The average signals of two biological experiments are shown. c Cells were grown and treated as in b. Rme1 binding to the IRT1 promoter was measured by chromatin immunoprecipitation at 4 h after treatment. Cells were fixed with formaldehyde, following immunoprecipitation of Rme1 tagged with 3xV5 epitope (FW1765) from chromatin extracts (see “Materials and methods” for details). The recovered DNA was quantified by real-time PCR with primers corresponding to the IRT1 promoter (pIRT1). Signals were normalized to the silent mating-type locus (HMR), which does not bind Rme1
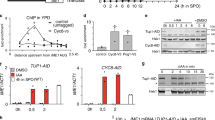
Northern blot analysis of IRT1 in wild-type and tup1Δ cells during diauxic shift. Haploid control (FW1509) or tup1Δ (FW2752) cells were grown overnight, diluted in YPD (OD600 = 2) and samples were taken at the indicated time points. Total RNA was isolated, separated by gel electrophoresis, blotted and probed for IRT1
Northern blot analysis
To measure IRT1 expression by northern blot we used a northern blot protocol which has been described previously (Koster et al. 2014). The IRT1 probe has been described previously (van Werven et al. 2012).
RT-PCR
The RT-PCR protocol was described previously (Weidberg et al. 2016). In short, 750 ng of DNase-treated total RNA was used for the reverse transcription reaction, and single-stranded cDNA were quantified by real-time PCR using an SYBR green mix (Life Technologies) on a 7500 Fast Real-Time PCR system (Life Technologies). Signals were normalized to ACT1 transcripts levels. The primer sequences are available on request.
Chromatin immunoprecipitation
Chromatin immunoprecipitation experiments were executed as described previously (Weidberg et al. 2016). Cells were fixed with 1 % formaldehyde for 20 min, the reaction was quenched with 125 mM glycine. Cells were disrupted using a mini beadbeater (BioSpec), and crosslinked chromatin was sheered by sonication using the Bioruptor (Diagenode, 7 cycles of 30 s on/off). Chromatin extracts were then incubated with anti-V5 agarose beads (Sigma) for 2 h at room temperature, and beads were washed accordingly. To measure Rme1 binding, input and ChIP samples were quantified by real-time PCR using SYBR green mix (Life Technologies) and primers corresponding to the IRT1 promoter on a 7500 Fast Real-Time PCR system (Life Technologies). The mating-type locus (HMR) was used as a non-binding negative control. Primer sequences are available on request.
Results and discussion
TORC1 and PKA control IRT1 transcription
Previous work showed that the TORC1 and PKA nutrient sensing pathways control the promoter of IME1, the master regulator of entry into sporulation (Weidberg et al. 2016). The IME1 promoter is not only regulated by nutrient availability, but also by mating type (Covitz and Mitchell 1993; Kassir et al. 1988; Mitchell and Herskowitz 1986). In cells with a single mating type, IME1 is repressed by transcription of the long non-coding RNA called IRT1, which transcribes through the IME1 promoter. Like IME1, induction of IRT1 occurs when haploid cells are starved (van Werven et al. 2012). It is unclear, however, how IRT1 stays repressed when nutrients are ample. We hypothesized that similar to IME1, IRT1 is controlled by TORC1 and PKA signalling. To test this, we used the analogue sensitive allele of PKA (tpk1as) that was described previously and the TORC1 inhibitor rapamycin (Weidberg et al. 2016). When we grew cells in nutrient-rich conditions (YPD) and inhibited PKA using the small molecule 1NM-PP1, IRT1 was strongly induced (Fig. 1a, b). Inhibition of TORC1 alone following rapamycin treatment had almost no effect on IRT1 transcription, but when combined with inhibition of PKA IRT1 levels increased further (Fig. 1a, b). We also measured the binding of Rme1, the activator of IRT1, to the IRT1 promoter (van Werven et al. 2012). We found that inhibition of PKA alone led to strong recruitment of Rme1, whereas inhibition of TORC1 resulted in low levels of Rme1 recruitment. The combined repression of PKA and TORC1 had little added effect when compared to PKA alone suggesting that PKA is the main regulator of IRT1 under these conditions. Similar observations were made for IME1 because inhibition of PKA has a much stronger effect on IME1 activation than inhibition of TORC1 (Weidberg et al. 2016). These results show that, like IME1, IRT1 transcription is under the control of the PKA and TORC1 signalling pathways.
Tup1 is required for IRT1 repression
Having established that TORC1 and PKA control IRT1 transcription in cells with a single mating type, we next examined whether IRT1 and IME1 also share the same repressor protein complex. In nutrient-rich conditions, the IME1 promoter is repressed by Tup1–Cyc8 complex, which binds in the middle of the IME1 promoter (between −800 and −1000 base pairs upstream to the transcription start site) (Mizuno et al. 1998; Weidberg et al. 2016). This transcriptional repressor interacts with sequence-specific transcription factors and histone de-acetyltransferases to repress promoters (Keleher et al. 1992; Tzamarias and Struhl 1994; Watson et al. 2000; Wong and Struhl 2011). Tup1–Cyc8 is evicted from the IME1 promoter to de-repress IME1, following nutrient starvation and inhibition of PKA/TORC1 (Weidberg et al. 2016). To test whether IRT1 is also regulated by Tup1–Cyc8, we measured IRT1 expression in the wild-type and tup1Δ cells in nutrient-rich conditions and post-diauxic shift when nutrients were largely used from the growth medium. We found that IRT1 was de-repressed in the tup1Δ cells exposed to high nutrients (OD600 ~ 2). Interestingly, the levels of IRT1 were significantly higher when tup1Δ cells were grown to high density (OD600 ~ 10). We conclude that Tup1–Cyc8 contributes to IRT1 repression.
Our result shows that, like IME1, IRT1 (at least in part) is regulated by Tup1–Cyc8. How Tup1–Cyc8 represses both IRT1 and IME1 remains unclear. Promoter scanning showed that Tup1 binding peaked in the middle of the IME1 promoter (Weidberg et al. 2016). We observed no difference in Tup1 binding between haploid and diploid cells (unpublished data). Given that the distance between the Rme1 and Tup1 binding in the IME1 promoter is relatively short (under one kilobase), we propose that Tup1–Cyc8 establishes a repressive chromatin state at the IME1 promoter that spreads to the IRT1 promoter. Our observation that IRT1 expression was further induced during post-diauxic shift in tup1Δ cells suggests that Tup1–Cyc8 is not the sole repressor of IRT1 and other factors may also contribute. Another explanation is that the activator of IRT1, Rme1, is regulated by nutrient availability. It is known that Rme1 levels are cell cycle regulated, peaking during late M/early G1 (Toone et al. 1995). Upon post-diauxic shift a large fraction of cells arrest in G1, whereas in nutrient-rich conditions cells continuously progress through the cell cycle. More work is needed to fully depict how IRT1 repression is controlled.
Conclusions and model
It has been known for decades that nutrient availability and mating type are key regulators of IME1 and entry into sporulation. How nutrients regulate mating-type control of IME1 was not understood. Here, we show that transcription of the mating-type-regulated lncRNA IRT1 is under the control of the same nutrient sensing pathways (TORC1 and PKA) as IME1. In addition, we provide evidence that IRT1 and IME1 are under control of the same repressor complex, Tup1–Cyc8. These findings have several implications. First, they show that mating-type control of IME1 is not active when nutrients are ample (Fig. 3). In other words, there is a hierarchy, in which nutrient repression is prevalent over mating-type control. Second, co-regulation of IME1 and IRT1 could serve as a fail-safe mechanism for mating-type control of IME1. Indeed, it has been known that mating-type control of sporulation is essential for preventing haploid cells to enter meiosis, which would be lethal as two consecutive cell divisions would attempt the segregation of a haploid genome into four spores. If IRT1 and IME1 were under control of different signalling pathways and different transcriptional repressors, this could have led to a mis-regulation of mating-type control of IME1. Our observation that IME1 and IRT1 expressions are regulated by the same nutrient sensing pathways and by the same transcriptional repressor complex ensures that IRT1 is activated at the same time when the nutrient requirements for IME1 are met and vice versa. Overall, we propose that the hierarchy between nutrient and mating-type signals ensures that diploid, and not haploid, cells induce IME1 and enter sporulation.
Model of nutrient and mating-type control of IME1. Under nutrient-rich conditions, TORC1/PKA repress IME1 and IRT1 via Tup1–Cyc8 in diploid/haploid cells harbouring either both or single mating types (MATa/α, MATa or MATα). During starvation, cells with a single mating type (MATa or MATα) induce IRT1-mediated repression of IME1, whereas MATa/α diploid cells induce IME1 and enter sporulation
References
Covitz PA, Mitchell AP (1993) Repression by the yeast meiotic inhibitor RME1. Genes Dev 7:1598–1608
Honigberg SM, Purnapatre K (2003) Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J Cell Sci 116:2137–2147. doi:10.1242/jcs.00460
Jin L, Neiman AM (2016) Post-transcriptional regulation in budding yeast meiosis. Curr Genet 62:313–315. doi:10.1007/s00294-015-0546-2
Kassir Y, Granot D, Simchen G (1988) IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 52:853–862
Kassir Y, Adir N, Boger-Nadjar E, Raviv NG, Rubin-Bejerano I, Sagee S, Shenhar G (2003) Transcriptional regulation of meiosis in budding yeast. Int Rev Cytol 224:111–171
Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD (1992) Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68:709–719
Koster MJ, Yildirim AD, Weil PA, Holstege FC, Timmers HT (2014) Suppression of intragenic transcription requires the MOT1 and NC2 regulators of TATA-binding protein. Nucleic Acids Res 42:4220–4229. doi:10.1093/nar/gkt1398
Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. doi:10.1002/(SICI)1097-0061(199807)14:10<953:AID-YEA293>3.0.CO;2-U
Mitchell AP, Herskowitz I (1986) Activation of meiosis and sporulation by repression of the RME1 product in yeast. Nature 319:738–742. doi:10.1038/319738a0
Mizuno T, Nakazawa N, Remgsamrarn P, Kunoh T, Oshima Y, Harashima S (1998) The Tup1-Ssn6 general repressor is involved in repression of IME1 encoding a transcriptional activator of meiosis in Saccharomyces cerevisiae. Curr Genet 33:239–247
Nachman I, Regev A, Ramanathan S (2007) Dissecting timing variability in yeast meiosis. Cell 131:544–556. doi:10.1016/j.cell.2007.09.044
Toone WM, Johnson AL, Banks GR, Toyn JH, Stuart D, Wittenberg C, Johnston LH (1995) Rme1, a negative regulator of meiosis, is also a positive activator of G1 cyclin gene expression. EMBO J 14:5824–5832
Tzamarias D, Struhl K (1994) Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature 369:758–761. doi:10.1038/369758a0
van Werven FJ, Amon A (2011) Regulation of entry into gametogenesis. Philos Trans R Soc Lond B Biol Sci 366:3521–3531. doi:10.1098/rstb.2011.0081
van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A (2012) Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell 150:1170–1181. doi:10.1016/j.cell.2012.06.049
Vershon AK, Pierce M (2000) Transcriptional regulation of meiosis in yeast. Curr Opin Cell Biol 12:334–339
Watson AD, Edmondson DG, Bone JR, Mukai Y, Yu Y, Du W, Stillman DJ, Roth SY (2000) Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev 14:2737–2744
Weidberg H, Moretto F, Spedale G, Amon A, van Werven FJ (2016) Nutrient control of yeast gametogenesis is mediated by TORC1, PKA and energy availability. PLoS Genet 12:e1006075. doi:10.1371/journal.pgen.1006075
Wong KH, Struhl K (2011) The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev 25:2525–2539. doi:10.1101/gad.179275.111
Acknowledgments
We thank the members of the van Werven lab and Hilla Weidberg for discussion and critical reading of the manuscript. The research of the van Werven lab is supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001203), the UK Medical Research Council (FC001203), and the Wellcome Trust (FC001203).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Moretto, F., van Werven, F.J. Transcription of the mating-type-regulated lncRNA IRT1 is governed by TORC1 and PKA. Curr Genet 63, 325–329 (2017). https://doi.org/10.1007/s00294-016-0639-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-016-0639-6