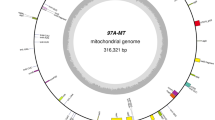
Abstract
Cytoplasmic male-sterility (CMS) conferred by the CMS-S cytoplasm has been most commonly used for onion (Allium cepa L.) F1 hybrid seed production. We first report the complete mitochondrial genome sequence containing CMS-S cytoplasm in this study. Initially, seven contigs were de novo assembled from 150-bp paired-end raw reads produced from the total genomic DNA using the Illumina NextSeq500 platform. These contigs were connected into a single circular genome consisting of 316,363 bp (GenBank accession: KU318712) by PCR amplification. Although all 24 core protein-coding genes were present, no ribosomal protein-coding genes, except rps12, were identified in the onion mitochondrial genome. Unusual trans-splicing of the cox2 gene was verified, and the cox1 gene was identified as part of the chimeric orf725 gene, which is a candidate gene responsible for inducing CMS. In addition to orf725, two small chimeric genes were identified, but no transcripts were detected for these two open reading frames. Thirteen chloroplast-derived sequences, with sizes of 126–13,986 bp, were identified in the intergenic regions. Almost 10 % of the onion mitochondrial genome was composed of repeat sequences. The vast majority of repeats were short repeats of <100 base pairs. Interestingly, the gene encoding ccmFN was split into two genes. The ccmF N gene split is first identified outside the Brassicaceae family. The breakpoint in the onion ccmF N gene was different from that of other Brassicaceae species. This split of the ccmF N gene was also present in 30 other Allium species. The complete onion mitochondrial genome sequence reported in this study would be fundamental information for elucidation of onion CMS evolution.





Similar content being viewed by others
References
Abdelnoor RV, Christensen AC, Mohammed S, Munoz-Castillo B, Moriyama H, Mackenzie SA (2006) Mitochondrial genome dynamics in plants and animals: convergent gene fusions of a MutS homologue. J Mol Evol 63:165–173
Adams KL, Qiu Y, Stoutemyer M, Palmer JD (2002) Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci 99:9905–9912
Albert B, Godelle B, Gouyon PH (1998) Evolution of the plant mitochondrial genome: dynamics of duplication and deletion of sequences. J Mol Evol 46:155–158
Allen JO, Fauron CM, Mink P, Roark L, Oddiraju S, Lin GN, Meyer L, Sun H, Kim K, Wang C, Du F, Xu D, Gibson M, Cifrese J, Clifton SW, Newton KJ (2007) Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics 177:1173–1192
Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD (2010) Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol 27:1436–1448
Alverson AJ, Rice DW, Dickinson S, Barry K, Palmer JD (2011a) Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell 23:2499–2513
Alverson AJ, Zhuo S, Rice DW, Sloan DB, Palmer JD (2011b) The mitochondrial genome of the legume Vigna radiata and the analysis of recombination across short mitochondrial repeats. PLoS One 6:e16404
Archibald JM (2015) Endosymbiosis and eukaryotic cell evolution. Curr Biol 25:R911–R921
Arrieta-Montiel M, Lyznik A, Woloszynska M, Janska H, Tohme J, Mackenzie SA (2001) Tracing evolutionary and developmental implications of mitochondrial stoichiometric shifting in the common bean. Genetics 158:851–864
Backert S, Neilsen BL, Börner T (1997) The mystery of the rings: structure and replication of mitochondrial genomes from higher plants. Trend Plant Sci 2:477–483
Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65:415–442
Bellaoui M, Martin-Canadell A, Pelletier G, Budar F (1998) Low-copy-number molecules are produced by recombination, actively maintained and can be amplified in the mitochondrial genome of Brassicaceae: relationship to reversion of the male sterile phenotype in some cybrids. Mol Gen Genet 257:177–185
Bentolila S, Alfonso AA, Hanson MR (2002) A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci 99:10887–10892
Berninger E (1965) Contribution à l’étude de la sterilité mâle de l’oignon (Allium cepa L.). Ann Amélior Plant 15:183–199
Brown GG, Formanova N, Jin H, Wargachuk R, Dendy C, Patil P, Laforest M, Zhang J, Cheung WY, Landry BS (2003) The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J 35:262–272
Budar F, Touzet P, De Paepe R (2003) The nucleo-mitochondrial conflict in cytoplasmic male sterilities revised. Genetica 117:3–16
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Conant GC, Wolfe KH (2008) GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics 24:861–862
Courcel AGL, de Vedel F, Boussac JM (1989) DNA polymorphism in Allium cepa cytoplasms and its implications concerning the origin of onions. Theor Appl Genet 77:793–798
Cui X, Wise RP, Schnable PS (1996) The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 272:1334–1336
Desloire S, Gherbi H, Laloui W, Marhadour S, Clouet V, Cattolico L, Falentin C, Giancola S, Renard M, Budar F, Small I, Caboche M, Delourme R, Bendahmane A (2003) Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep 4:588–594
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M (2014) Pfam: the protein families database. Nucl Acids Res 42:D222–D230
Fujii S, Toriyama K (2009) Suppressed expression of retrograde-regulated male sterility restores pollen fertility in cytoplasmic male sterile rice plants. Proc Natl Acad Sci 106:9513–9518
Grewe F, Edger PP, Keren I, Sultan L, Pires JC, Ostersetzer-Biran O, Mower JP (2014) Comparative analysis of 11 Brassicales mitochondrial genomes and the mitochondrial transcriptome of Brassica oleracea. Mitochondrion 19:135–143
Guo W, Grewe F, Fan W, Young GJ, Knoop V, Palmer JD, Mower JP (2016) Ginkgo and Welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol Biol Evol (in press)
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Window 95/98/NT. Nucl Acids Symp Ser 41:95–98
Handa H (2003) The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucl Acids Res 31:5907–5916
Hanson MR, Bentolila S (2004) Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16:S154–S169
Havey MJ (1993) A putative donor of S-cytoplasm and its distribution among open-pollinated populations of onion. Theor Appl Genet 86:128–134
Hecht J, Grewe F, Knoop V (2011) Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol 3:344–358
Holford P, Croft JH, Newbury HJ (1991) Differences between, and possible origins of, the cytoplasms found in fertile and male-sterile onions (Allium cepa L.). Theor Appl Genet 82:737–744
Itabashi E, Iwata N, Fujii S, Kazama T, Toriyama K (2011) The fertility restorer gene, Rf2, for lead rice-type cytoplasmic male sterility of rice encodes a mitochondrial glycine-rice protein. Plant J 65:359–367
Janska H, Sarria R, Woloszynska M, Arrieta-Montiel M, Mackenzie SA (1998) Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. Plant Cell 10:1163–1180
Jones HA, Clarke A (1943) Inheritance of male sterility in the onion and the production of hybrid seed. Proc Am Soc Hortic Sci 43:189–194
Jones HA, Emsweller SL (1936) A male-sterile onion. Proc Am Soc Hortic Sci 34:582–585
Kim S (2013) Identification of hypervariable chloroplast intergenic sequences in onion (Allium cepa L.) and their use to analyse the origins of male-sterile onion cytotypes. J Hortic Sci Biotech 88:187–194
Kim S (2014) A codominant molecular marker in linkage disequilibrium with a restorer-of-fertility gene (Ms) and its application in reevaluation of inheritance of fertility restoration in onions. Mol Breed 34:769–778
Kim S, Yoon M (2010) Comparison of mitochondrial and chloroplast genome segments from three onion (Allium cepa L.) cytoplasm types and identification of a trans-splicing intron of cox2. Curr Genet 56:177–188
Kim S, Lim H, Park S, Cho K, Sung S, Oh D, Kim K (2007) Identification of a novel mitochondrial genome type and development of molecular makers for cytoplasm classification in radish (Raphanus sativus L.). Theor Appl Genet 115:1137–1145
Kim S, Lee E, Cho DY, Han T, Bang H, Patil BS, Ahn YK, Yoon M (2009) Identification of a novel chimeric gene, orf725, and its use in development of a molecular marker for distinguishing three cytoplasm types in onion (Allium cepa L.). Theor Appl Genet 118:433–441
Kim S, Bang H, Patil BS (2013) Origin of three characteristic onion (Allium cepa L.) mitochondrial genome rearrangements in Allium species. Sci Hortic 157:24–31
Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ (2015a) Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One 10:e0117159
Kim S, Kim C, Park M, Choi D (2015b) Identification of candidate genes associated with fertility restoration of cytoplasmic male-sterility in onion (Allium cepa L.) using a combination of bulked segregant analysis and RNA-seq. Theor Appl Genet 128:2289–2299
Kim S, Kim M, Kim Y, Yeom S, Cheong K, Kim K, Jeon J, Kim S, Kim D, Sohn S, Lee Y, Choi D (2015c) Integrative structural annotation of de novo RNA-Seq provides an accurate reference gene set of the enormous genome of the onion (Allium cepa L.). DNA Res 22:19–27
Kim S, Park J, Yang T (2015d) Comparative analysis of the complete chloroplast genome sequences of a normal male-fertile cytoplasm and two different cytoplasms conferring cytoplasmic male sterility in onion (Allium cepa L.). J Hortic Sci Biotech 90:459–468
Klauer AA, van Hoof A (2012) Degradation of mRNAs that lack a stop codon: a decade of nonstop progress. Wiley Interdiscip Rev RNA 3:649–660
Kmiec B, Woloszynska M, Janska H (2006) Heteroplasmy as a common state of mitochondrial genetic information in plants and animals. Curr Genet 50:149–159
Knoop V (2004) The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr Genet 46:123–139
Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, Sakai T, Kawasaki S, Imamura J (2003) Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J 34:407–415
Komori T, Ohta S, Murai N, Takakura Y, Kuraya Y, Suzuki S, Hiei Y, Imaseki H, Nitta N (2004) Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.). Plant J 37:315–325
Kubo T, Newton KJ (2008) Angiosperm mitochondrial genomes and mutations. Mitochondrion 8:5–14
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL (2004) Versatile and open software for comparing large genomes. Genome Biol 5:R12
Laser KD, Lersten NR (1972) Anatomy and cytology of microsporogenesis in cytoplasmic male sterile angiosperms. Bot Rev 38:425–454
Lohse M, Drechsel O, Kahlau S, Bock R (2013) OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucl Acids Res 41:W575–W581
Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Ohyama K (1992) Transfer RNA genes in the mitochondrial genome from a liverwort, Marchantia polymorpha: the absence of chloroplast-like tRNAs. Nucl Acids Res 20:3773–3777
Oldenburg DJ, Bendich AJ (2001) Mitochondrial DNA from the Liverwort Marchantia polymorpha: circularly permuted linear molecules, head-to-tail concatemers, and a 5′ protein. J Mol Biol 310:549–562
Palmer JD (1988) Intraspecific variation and multicircularity in Brassica mitochondrial DNAs. Genetics 118:341–351
Park JY, Lee Y, Lee J, Choi B, Kim S, Yang T (2013) Complete mitochondrial genome sequence and identification of a candidate gene responsible for cytoplasmic male sterility in radish (Raphanus sativus L.) containing DCGMS cytoplasm. Theor Appl Genet 126:1763–1774
Raczynska KD, Le Ret M, Rurek M, Bonnard G, Augustyniak H, Gualberto JM (2006) Plant mitochondrial genes can be expressed from mRNAs lacking stop codons. FEBS Lett 580:5641–5646
Rayapuram N, Hagenmuller J, Grienenberger JM, Bonnard G, Giegé P (2008) The three mitochondrial encoded ccmF proteins form a complex that interacts with CCMH and c-type apocytochromes in Arabidopsis. J Biol Chem 283:25200–25208
Rice DW, Alverson AJ, Richardson AO, Young GJ, Sanchez-Puerta MV, Munzinger J, Barry K, Boore JL, Zhang Y, dePamphilis CW, Knox EB, Palmer JD (2013) Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 342:1468–1473
Sakai T, Imamura J (1993) Evidence for a mitochondrial sub-genome containing radish AtpA in a Brassica napus cybrid. Plant Sci 90:95–103
Sandhu AP, Abdelnoor RV, Mackenzie SA (2007) Transgenic induction of mitochondrial rearrangements for cytoplasmic male sterility in crop plants. Proc Natl Acad Sci 104:1766–1770
Schnable PS, Wise RP (1998) The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci 3:175–180
Schweisguth B (1973) Étude d’un nouveau type de stérilité male chez l’oignon, Allium cepa L. Ann Amélior Plant 23:221–233
Shedge V, Arrieta-Montiel M, Christensen AC, Mackenzie SA (2007) Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell 19:1251–1264
Skippington E, Barkman TJ, Rice DW, Palmer JD (2015) Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc Natl Acad Sci 112:E3515–E3524
Sloan DB (2013) One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol 200:978–985
Sloan DB, Alverson AJ, Chuckalovcak JP, Wu M, McCauley DE, Palmer JD, Taylor DR (2012a) Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol 10:e1001241
Sloan DB, Müller K, McCauley DE, Taylor DR, Štorchová H (2012b) Intraspecific variation in mitochondrial genome sequence, structure, and gene content in Silene vulgaris, an angiosperm with pervasive cytoplasmic male sterility. New Phytol 196:1228–1239
Small I, Suffolk R, Leaver CJ (1989) Evolution of plant mitochondrial genomes via substoichiometric intermediates. Cell 58:69–76
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tanaka Y, Tsuda M, Yasumoto K, Terachi T, Yamagishi H (2014) The complete mitochondrial genome sequence of Brassica oleracea and analysis of coexisting mitotypes. Curr Genet 60:277–284
Thöny-Meyer L (2002) Cytochrome c maturation: a complex pathway for a simple task? Biochem Soc Trans 30:633–638
Unseld M, Marienfeld JR, Brandt P, Brennicke A (1997) The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet 15:57–61
Wang W, Wu Y, Messing J (2012) The mitochondrial genome of an aquatic plant, Spirodela polyrhiza. PLoS One 7:e46747
Woloszynska M, Trojanowski D (2009) Counting mtDNA molecules in Phaseolus vulgaaris: sublimons are constantly produced by recombination via short repeats and undergo rigorous selection during substoichiometric shifting. Plant Mol Biol 70:511–521
Wu Z, Cuthberta JM, Taylorb DR, Sloana DB (2015) The massive mitochondrial genome of the angiosperm Silene noctiflora is evolving by gain or loss of entire chromosomes. Proc Natl Acad Sci 112:10185–10191
Zaegel V, Guermann B, Le Ret M, Andrés C, Meyer D, Erhardt M, Canaday J, Gualberto JM, Imbault P (2006) The plant-specific ssDNA binding protein OSB1 is involved in the stoichiometric transmission of mitochondrial DNA in Arabidopsis. Plant Cell 18:3548–3563
Zhang T, Zhang X, Hu S, Yu J (2011) An efficient procedure for plant organellar genome assembly, based on whole genome data from the 454 GS FLX sequencing platform. Plant Methods 7:38
Acknowledgments
This study was supported by the Agriculture Research Center program, Golden Seed Project (Center for Horticultural Seed Development, No 213003-04-2-SB910) and a Grant from the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center No. PJ011034). The authors thank Ji-wha Hur, Jeong-Ahn Yoo, and Su-jung Kim for their dedicated technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Communicated by M. Kupiec.
Electronic supplementary material
Below is the link to the electronic supplementary material.
294_2016_595_MOESM1_ESM.tif
Supplementary Fig. 1. Organization of seven contigs assembled using sequence reads produced by next-generation sequencing. Genes transcribed as forward and reverse complements are indicated as boxes on and beneath the lines, respectively. The filled and empty boxes indicate exons and introns, respectively (TIFF 97 kb)
294_2016_595_MOESM2_ESM.tif
Supplementary Fig. 2. Positions of the seven contigs on the onion mitochondrial genome. A. Onion mitochondrial genome map showing positions of the seven contigs. Bars on the contigs indicate positions of the polymerase chain reaction (PCR) products amplified using primers binding to the end sequences of each contig or chloroplast-derived sequences. B. PCR products amplified using primer pairs binding to the junctions of the contigs. The primer sequences are shown in Supplementary Table 2. Con: PCR product amplified using a primer pair binding to the closely linked nad3 and rps12 genes, respectively (TIFF 334 kb)
294_2016_595_MOESM3_ESM.tif
Supplementary Fig. 3. Structure of three chimeric genes identified in the onion mitochondrial genome. Arrow-shaped boxes indicate the 5′-to-3′ direction. Empty and gray boxes indicate exons and uncharacterized sequences, respectively (TIFF 54 kb)
294_2016_595_MOESM4_ESM.tif
Supplementary Fig. 4. Onion mitochondrial genome map showing the positions and sizes of chloroplast-derived sequences. The detail information of these 13 fragments is shown in Supplementary Table 3 (TIFF 56 kb)
294_2016_595_MOESM5_ESM.tif
Supplementary Fig. 5. Assessment of recombination frequency of two large-sized repeats. A. Diagram showing the positions of primers used to detect normal and recombinant conformations. B. PCR amplification of normal and recombinant conformations. Detail information about the R1 and R2 repeats are shown in Supplementary Table 7. PCR was performed with a 35-cycle condition and primer sequences are shown in Supplementary Table 2 (TIFF 211 kb)
Rights and permissions
About this article
Cite this article
Kim, B., Kim, K., Yang, TJ. et al. Completion of the mitochondrial genome sequence of onion (Allium cepa L.) containing the CMS-S male-sterile cytoplasm and identification of an independent event of the ccmF N gene split. Curr Genet 62, 873–885 (2016). https://doi.org/10.1007/s00294-016-0595-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-016-0595-1