Abstract
Among other approaches, differential equations are used for a deterministic quantitative description of time-dependent biological processes. For intracellular systems, such as signaling pathways, most existing models are based on ordinary differential equations. These models describe temporal processes, while they neglect spatial aspects. We present a model for the SMAD signaling pathway, which gives a temporal and spatial description on the basis of reaction diffusion equations to answer the question whether cell geometry plays a role in signaling. In this article we simulate the ordinary differential equations as well as partial differential equations of parabolic type with suile numerical methods, the latter on different cell geometries. In addition to manual construction of idealized cells, we also construct meshes from microscopy images of real cells. The main focus of the paper is to compare the results of the model without and with spatial aspects to answer the addressed question. The results show that diffusion in the model can lead to significant intracellular gradients of signaling molecules and changes the level of response to the signal transduced by the signaling pathway. In particular, the extent of these observations depends on the geometry of the cell.














Similar content being viewed by others
References
ANSYS ICEM CFD (2010) Mesh generation. http://www.ansys.com/products/icemcfd.asp
Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, Ryan S, Spiller DG, Unitt JF, Broomhead DS, Kell DK, Rand DA, Se V, White MRH (2009) Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science 324(5924):242–246
Brown GC, Kholodenko BN (1999) Spatial gradients of cellular phospho-proteins. FEBS Lett 457:452–454
Caudron M, Bunt G, Bastiaens P, Karsenti E (2005) Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science 309(5739):1373–1376
Chung S-W, Miles FL, Sikes RA, Cooper CR, Farach-Carson MC, Ogunnaike BA (2009) Quantitative modeling and analysis of the transforming growth factor signaling pathway. Biophys J 96(5):1733–1750
Ciarlet PG (1987) The finite element method for elliptic problems. North-Holland, Dordrecht
Clarke DC, Liu X (2008) Decoding the quantitative nature of TGF-\(\beta \)/Smad signaling. Trends Cell Biol 18(9):430–442
Claus J (2010) Spatial aspects in the simulation of the SMAD signal transduction pathway. Diploma thesis, Universität Heidelberg, Institut für Wissenschaftliches Rechnen
Deheuninck J, Luo K (2009) Ski and SnoN, potent negative regulators of TGF-beta signaling. Cell Res 19(1):47–57
GASCOIGNE (2002) High performance adaptive finite element toolkit. http://www.numerik.uni-kiel.de/mabr/gascoigne/
Gonzlez-Prez V, Schmierer B, Hill CS, Sear RP (2011) Studying SMAD2 intranuclear diffusion dynamics by mathematical modelling of FRAP experiments. Integr Biol (Camb) 3(3):197–207
Heldin C-H, Miyazono K, ten Dijke P (1997) TGF-\(\beta \) signalling from cell membrane to nucleus through SMAD proteins. Nature 390:465–471
Hengl S, Kreutz C, Timmer J, Maiwald T (2007) Data-based identifiability analysis of non-linear dynamical models. Bioinformatics 23(19):2612–2618
Jungblut D (2010) Rekonstruktion von Oberflächenmorphologien und Merkmalskeletten aus dreidimensionalen Daten unter Verwendung hochparalleler Rechnerarchitekturen. PhD thesis, Goethe-Universität Frankfurt am Main
Kajino T, Omori E, Ishii S, Ninomiya-Tsuji J (2007) TAK1 MAPK kinase kinase mediates transforming growth factor-beta signaling by targeting SnoN oncoprotein for degradation. J Biol Chem 282(13):9475–9481
Kholodenko BN, Brown GC, Hoek JB (2000) Diffusion control of protein phosphorylation in signal transduction pathways. Biochem J 350:901–907
Kholodenko BN (2006) Cell signalling dynamics in time and space. Nat Rev Mol Cell Biol 7(3):165–176
Klingmüller U, Bauer A, Bohl S, Nickel PJ, Breitkopf K, Dooley S, Zellmer S, Kern C, Merfort I, Sparna T, Donauer J, Walz G, Geyer M, Kreutz C, Hermes M, Götschel F, Hecht A, Walter D, Egger L, Neubert K, Borner C, Brulport M, Schormann W, Sauer C, Baumann F, Preiss R, MacNelly S, Godoy P, Wiercinska E, Ciuclan L, Edelmann J, Zeilinger K, Heinrich M, Zanger UM, Gebhardt R, Maiwald T, Heinrich R, Timmer J, von Weizscker F, Hengstler JG (2006) Primary mouse hepatocytes for systems biology approaches: a standardized in vitro system for modelling of signal transduction pathways. Syst Biol (Stevenage) 153:433–447
Legewie S (2009) Systems biological analyses of intracellular signal transduction. PhD thesis, Humboldt-Universität Berlin, Mathematisch-Naturwissenschaftliche Fakultät I
Lev Bar-Or R, Maya R, Segel LA, Alon U, Levine AJ, Oren M (2000) Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. PNAS
Melke P, Jönsson H, Pardali E, ten Dijke P, Peterson C (2006) A rate equation approach to elucidate the kinetics and robustness of the TGF-pathway. Biophys J 91(12):4368–4380
Meyers J, Craig J, Odde DJ (2006) Potential for control of signaling pathways via cell size and shape. Curr Biol 16(17):1685–1693
Neumann R (2009) Räumliche Aspekte der Signaltransduktion. Diploma thesis, Universität Heidelberg, Institut für wissenschaftliches Rechnen
NeuRA2 (2009) The neuron reconstruction algorithm. http://www.neura.org
Neves SR, Tsokas P, Sarkar A, Grace EA, Rangamani P, Taubenfeld SM, Alberini CM, Schaff JC, Blitzer RD, Moraru II, Iyengar R (2008) Cell shape and negative links in regulatory motifs together control spatial information flow in signaling networks. Cell 133(4):666–680
Schmierer B, Tournier AL, Bates PA, Hill CS (2008) Mathematical modeling identifies Smad nucleocytoplasmic shuttling as a dynamic signal-interpreting system. Proc Natl Acad Sci USA 105(18):6608–6613
Straube R, Ward MJ (2009) An asymptotic analysis of intracellular signaling gradients arising from multiple small compartments. SIAM J Appl Math 70:248–269
Tewari M, Rao A (2006) Systems biology approaches to TGF-/Smad signaling. In: Dijke P, ten Heldin C-H, Ridley A, Frampton J (eds) Proteins and cell regulation, Smad signal transduction, vol 5. Springer, Berlin, pp 361–378
Wilkinson DS, Ogden SK, Stratton SA, Piechan JL, Nguyen TT, Smulian GA, Barton MC (2005) A direct intersection between p53 and transforming growth factor beta pathways targets chromatin modification and transcription repression of the alpha-fetoprotein gene. Mol Cell Biol 25(3):1200–1212
Yang J, Dai C, Liu Y (2005) A novel mechanism by which hepatocyte growth factor blocks tubular epithelial to mesenchymal transition. J Am Soc Nephrol 16(1):68–78
Zi Z, Klipp E (2007) Constraint-based modeling and kinetic analysis of the Smad dependent TGF-signaling pathway. PLoS ONE 2(9):e936
Zvaifler NJ (2006) Relevance of the stroma and epithelial-mesenchymal transition EMT for the rheumatic diseases. Arthrit Res Therapy 8:210
Acknowledgments
Special thanks to Dr. D. Jungblut, Goethe Center for Scientific Computing (G-CSC), University Frankfurt, Germany, for doing the 3D geometry reconstruction with his software developed in his PhD.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work was supported by the Helmholtz Alliance on Systems Biology (SB Cancer, Submodule V.7).
Appendix: Experimental procedures
Appendix: Experimental procedures
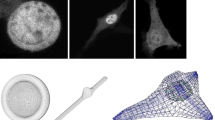
Isolated primary mouse hepatocytes (Klingmüller et al. 2006) were seeded at a density of \(4\times 10^4\) cells per well in collagen-coated 2-well Lab-Tek Chamber Slides. On the following day, cells were transiently transfected with GFP-SMAD2 expression plasmid with Lipofectamine 2000 reagent according to manufacturers protocol. After 16 hours of incubation, cells were stained for 15 min in 1:10,000 Hoechst 34580. Unbound dyes were removed by washing cells 3 times with cultivation medium. For reconstruction, imaging was performed (see Fig. 15) under cell culture conditions on a Zeiss 710 confocal microscope equipped with incubation chamber. Using a 63\(\times \)/1.40 Oil objective, Z-stack method in a 2-channel acquisition mode was applied: Hoechst-nucleus (excitation at 405 nm, and detection at 435–475 nm) and GFP-cytoplasm (excitation at 488 nm, and detection at 495–550 nm). The resulted voxel dimension is \(x:y:z = 0.264\times 0.264\times 0.364~\mu \)m\(^3\).
Rights and permissions
About this article
Cite this article
Claus, J., Friedmann, E., Klingmüller, U. et al. Spatial aspects in the SMAD signaling pathway. J. Math. Biol. 67, 1171–1197 (2013). https://doi.org/10.1007/s00285-012-0574-1
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-012-0574-1