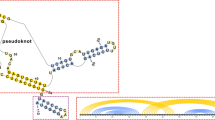
Abstract
Let \({\mathcal {S}}\) denote the set of (possibly noncanonical) base pairs {i, j} of an RNA tertiary structure; i.e. \({\{i, j\} \in \mathcal {S}}\) if there is a hydrogen bond between the ith and jth nucleotide. The page number of \({\mathcal {S}}\), denoted \({\pi(\mathcal {S})}\), is the minimum number k such that \({\mathcal {S}}\) can be decomposed into a disjoint union of k secondary structures. Here, we show that computing the page number is NP-complete; we describe an exact computation of page number, using constraint programming, and determine the page number of a collection of RNA tertiary structures, for which the topological genus is known. We describe an approximation algorithm from which it follows that \({\omega(\mathcal {S}) \leq \pi(\mathcal {S}) \leq \omega(\mathcal {S}) \cdot \log n}\), where the clique number of \({\mathcal {S}, \omega(\mathcal {S})}\), denotes the maximum number of base pairs that pairwise cross each other.
Similar content being viewed by others
References
Abrahams JP, van den Berg M, van Batenburg E, Pleij C (1990) Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucleic Acids Res 18: 3035–3044
Bellaousov S, Mathews DH (2010) Probknot: fast prediction of RNA secondary structure including pseudoknots. RNA 16(10): 1870–1880
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28(1): 235–242
Bon M (2009) Prédiction de structures secondaires d’ARN avec pseudo-noeuds. PhD thesis, Ecole Polytechnique
Bon M, Orland H (2011) TT2NE: a novel algorithm to predict RNA secondary structures with pseudoknots. Nucleic Acids Res 39(14): e93
Bon M, Vernizzi G, Orland H, Zee A (2008) Topological classification of RNA structures. J Mol Biol 379(4): 900–911
Cao S, Chen SJ (2006) Predicting RNA pseudoknot folding thermodynamics. Nucleic Acids Res 34(9): 2634–2652
Chen WY, Han HS, Reidys CM (2009) Random K-noncrossing RNA structures. Proc Natl Acad Sci USA 106(52): 22061–22066
Chung FRK, Leighton FT, Rosenberg AL (1987) Embedding graphs in books: a layout problem with applications to VLSI design. SIAM J Algebraic Discrete Methods 8(1): 33–58
Deigan KE, Li TW, Mathews DH, Weeks KM (2009) Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci USA 106(1): 97–102
Dirks RM, Pierce NA (2003) A partition function algorithm for nucleic acid secondary structure including pseudoknots. J Comput Chem 24(13): 1664–1677
Filotti IS, Miller GL, Reif JH (1979) On determining the genus of a graph in O(νO(g)) steps. In: STOC. ACM, pp 27–37
Gardner PP, Giegerich R (2004) A comprehensive comparison of comparative RNA structure prediction approaches. BMC Bioinformatics 5: 140
Garey MR, Johnson DS (1990) Computers and intractability: a guide to the theory of NP-completeness. W.H. Freeman & Co., New York
Garey MR, Johnson DS, Miller GL, Papadimitriou CH (1980) The complexity of coloring circular arcs and chords. SIAM J Algebraic Discrete Methods 1: 216–227
Golumbic MC (2004) Algorithmic graph theory and perfect graphs. North-Holland
Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR (2003) Rfam: an RNA family database. Nucleic Acids Res 31(1): 439–441
Gutell R, Lee J, Cannone J (2005) The accuracy of ribosomal RNA comparative structure models. Curr Opin Struct Biol 12: 301–310
Harary F (1994) Graph theory. Addison-Wesley, Reading
Haslinger C, Stadler PF (1999) Rna structures with pseudo-knots: graph-theoretical, combinatorial, and statistical properties. Bull Math Biol 61(3): 437–467
Heath LS, Istrail S (1987) The pagenumber of genus g graphs is O(g). In: STOC ’87: proceedings of the 19th annual ACM symposium on theory of computing, New York, NY, USA. ACM, pp 388–397
Hofacker I (2003) Vienna RNA secondary structure server. Nucleic Acids Res 31(13): 3429–3431
Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P (1994) Fast folding and comparison of RNA secondary structures. Monatsch Chem 125: 167–188
Huang Z, Wu Y, Robertson J, Feng L, Malmberg RL, Cai L (2008) Fast and accurate search for non-coding RNA pseudoknot structures in genomes. Bioinformatics 24(20): 2281–2287
Huang FW, Peng WW, Reidys CM (2009) Folding 3-noncrossing RNA pseudoknot structures. J Comput Biol 16(11): 1549–1575
Jensen TR, Toft B (1995) Graph coloring problems. Wiley, New York
Jin EY, Reidys CM (2010) On the decomposition of k-noncrossing RNA structures. Adv Appl Math 44(1): 53–70
Karapetyan IA (1980) Coloring of arc graphs. Akad Nauk Armyam SSR Doklady 70: 306–311 (in Russian)
Knudsen B, Hein J (2003) Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res 31(13): 3423–3428
Kostochka A, Kratochvil J (1997) Covering and coloring polygon-circle graphs. Discrete Math 163(1): 299–305
Leontis NB, Westhof E (2001) Geometric nomenclature and classification of RNA base pairs. RNA 7(4): 499–512
Lowe T, Eddy S (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5): 955–964
Lyngso RB, Pedersen CN (2000) RNA pseudoknot prediction in energy-based models. J Comput Biol 7(3-4): 409–427
Markham NR, Zuker M (2008) UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol 453: 3–31
Mathews DH, Sabina J, Zuker M, Turner H (1999) Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J Mol Biol 288: 911–940
Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH (2004) Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA 101: 7287–7292
Metzler D, Nebel ME (2008) Predicting RNA secondary structures with pseudoknots by MCMC sampling. J Math Biol 56(1–2): 161–181
Meyer IM, Miklos I (2007) Simulfold: simultaneously inferring RNA structures including pseudoknots, alignments, and trees using a Bayesian MCMC framework. PLoS Comput Biol 3(8): e149
Micali S, Vazirani VV (1980) An \({O (\sqrt{|V|} |E|)}\) algorithm for finding maximum matching in general graphs. In: 21st Annual symposium on foundations of computer science, pp 17–27
Poolsap U, Kato Y, Akutsu T (2009) Prediction of RNA secondary structure with pseudoknots using integer programming. BMC Bioinformatics 10: S38
Reeder J, Giegerich R (2004) Design, implementation and evaluation of a practical pseudoknot folding algorithm based on thermodynamics. BMC Bioinformatics 5: 104
Reidys CM, Huang FW, Andersen JE, Penner RC, Stadler PF, Nebel ME (2011) Topology and prediction of RNA pseudoknots. Bioinformatics 27(8): 1076–1085
Ren J, Rastegari B, Condon A, Hoos HH (2005) Hotknots: heuristic prediction of RNA secondary structures including pseudoknots. RNA 11(10): 1494–1504
Rivas E, Eddy SR (1999) A dynamic programming algorithm for RNA structure prediction including pseudoknots. J Mol Biol 285: 2053–2068
Sato K, Morita K, Sakakibara Y (2008) PSSMTS: position specific scoring matrices on tree structures. J Math Biol 56(1–2): 201–214
Sato K, Kato Y, Hamada M, Akutsu T, Asai K (2011) IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming. Bioinformatics 27(13): i85–i93
Smit S, Rother K, Heringa J, Knight R (2008) From knotted to nested RNA structures: a variety of computational methods for pseudoknot removal. RNA 14(3): 410–416
Sussman JL, Holbrook SR, Warrant RW, Church GM, Kim SH (1978) Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J Mol Biol 123(4): 607–630
Tabaska JE, Cary RE, Gabow HN, Stormo GD (1998) An RNA folding method capable of identifying pseudoknots and base triples. Bioinformatics 14: 691–699
Taufer M, Licon A, Araiza R, Mireles D, Van Batenburg FH, Gultyaev AP, Leung MY (2009) Pseudobase++: an extension of PseudoBase for easy searching, formatting and visualization of pseudoknots. Nucleic Acids Res 37(Database): D127–D135
Theimer CA, Giedroc DP (1999) Equilibrium unfolding pathway of an H-type RNA pseudoknot which promotes programmed-1 ribosomal frameshifting. J Mol Biol 289(5): 1283–1299
Thomassen C (1989) The graph genus problem is NP-complete. J Algorithms 10(4): 568–576
Van Batenburg FH, Gultyaev AP, Pleij CW (2001) Pseudobase: structural information on RNA pseudoknots. Nucleic Acids Res 29(1): 194–195
Van Hentenryck P (1989) Constraint satisfaction in logic programming. The MIT Press, Cambridge
Vendruscolo M, Kussell E, Domany E (1997) Recovery of protein structure from contact maps. Fold Des 2: 295–306
Vernizzi G, Orland H, Zee A (2005) Enumeration of RNA structures by matrix models. Phys Rev Lett 94(16): 168103
Vernizzi G, Ribeca P, Orland H, Zee A (2006) Topology of pseudoknotted homopolymers. Phys Rev E 73(3): 031902
Wiese KC, Glen E, Vasudevan A (2005) JViz.Rna—a Java tool for RNA secondary structure visualization. IEEE Trans Nanobioscience 4(3): 212–218
Yang H, Jossinet F, Leontis N, Chen L, Westbrook J, Berman HM, Westhof E (2003) Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res 31(13): 3450–3560
Zhao J, Malmberg RL, Cai L (2008) Rapid ab initio prediction of RNA pseudoknots via graph tree decomposition. J Math Biol 56(1–2): 145–159
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31(13): 3406–3415
Zuker M, Stiegler P (1981) Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res 9(1): 133–148
Author information
Authors and Affiliations
Corresponding author
Additional information
P. Clote and I. Dotu: Research supported in part by National Science Foundation NSF grants DMS-1016618 and DMS-0817971, and by Digiteo Foundation; S. Dobrev: Supported in part by VEGA and APVV grants; and E. Kranakis: Research supported in part by NSERC and MITACS grants.
Rights and permissions
About this article
Cite this article
Clote, P., Dobrev, S., Dotu, I. et al. On the page number of RNA secondary structures with pseudoknots. J. Math. Biol. 65, 1337–1357 (2012). https://doi.org/10.1007/s00285-011-0493-6
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-011-0493-6