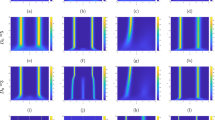
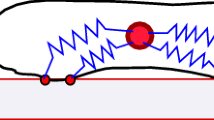
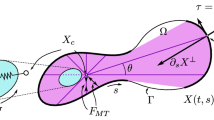
Abstract
The motility of cells crawling on a substratum has its origin in a thin cell organ called lamella. We present a 2-dimensional continuum model for the lamella dynamics of a slowly migrating cell, such as a human keratinocyte. The central components of the model are the dynamics of a viscous cytoskeleton capable to produce contractile and swelling stresses, and the formation of adhesive bonds in the plasma cell membrane between the lamella cytoskeleton and adhesion sites at the substratum. We will demonstrate that a simple mechanistic model, neglecting the complicated signaling pathways and regulation processes of a living cell, is able to capture the most prominent aspects of the lamella dynamics, such as quasi-periodic protrusions and retractions of the moving tip, retrograde flow of the cytoskeleton and the related accumulation of focal adhesion complexes in the leading edge of a migrating cell. The developed modeling framework consists of a nonlinearly coupled system of hyperbolic, parabolic and ordinary differential equations for the various molecular concentrations, two elliptic equations for cytoskeleton velocity and hydrodynamic pressure in a highly viscous two-phase flow, with appropriate boundary conditions including equalities and inequalities at the moving boundary. In order to analyse this hybrid continuum model by numerical simulations for different biophysical scenarios, we use suitable finite element and finite volume schemes on a fixed triangulation in combination with an adaptive level set method describing the free boundary dynamics.
Similar content being viewed by others
References
Adalsteinsson D., Sethian J.: The fast construction of extension velocities in level set methods. J. Comput. Phys. 148(1), 2–22 (1999)
Alt W.: Biomechanics of actomyosin-dependent mobility of keratinocytes. Biophysics 41(1), 181 (1996)
Alt W., Dembo M.: Cytoplasm dynamics and cell motion: two-phase flow models. Math. Biosci. 156, 207–228 (1999)
Alt W., Tranquillo R.T.: Protrusion-retraction dynamics of an annular lamellipodial seam. In: Alt, W. et al. (eds) Dynamics of Cell and Tissue Motion, pp. 73–81. Birkhäuser, Basel (1997)
Balaban N., Schwarz U., Riveline D., Goichberg P., Tzur G., Sabanay I., Mahalu D., Safran S., Bershadsky A., Addadi L., Geiger B.: Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nature Cell Biol. 3, 466–472 (2001)
Beningo K., Dembo M., Kaverina I., Small J., Wang Y.: Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 153(4), 881–888 (2001)
Bertolazzi E., Manzini G.: A cell-centered second-order accurate finite volume method for convection-diffusion problems on unstructured meshes. Math. Models Methods Appl. Sci 14(8), 1235–1260 (2004)
Borm B., Requardt R., Herzog V., Kirfel G.: Membrane ruffles in cell migration: indicators of inefficient lamellipodia adhesion and compartments of actin filament reorganization. Exp. Cell Res. 302(1), 83–95 (2005)
Bottino D., Mogilner A., Roberts T., Stewart M., Oster G.: How nematode sperm crawl. J. Cell Sci. 115(2), 367–384 (2002)
Brenner S., Scott L.: The Mathematical Theory of Finite Element Methods. Springer, Heidelberg (2002)
Bretschneider T., Diez S., Anderson K., Heuser J., Clarke M., Müller-Taubenberger A., Köhler J., Gerisch G.: Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr. Biol. 14(1), 1–10 (2004)
Bruinsma R.: Theory of force regulation by nascent adhesion sites. Biophys. J. 89(1), 87–94 (2005)
Burridge K., Chrzanowska-Wodnicka M., Zhong C.: Focal adhesion assembly. Trends Cell Biol. 7(9), 342–347 (1997)
Chung C., Funamoto S., Firtel R.: Signaling pathways controlling cell polarity and chemotaxis. Trends Biochem. Sci. 26(9), 557–566 (2001)
Dembo M.: Mechanics and control of the cytoskeleton in Amoeba proteus. Biophys. J. 55(6), 1053–1080 (1989)
Frank D., Carter W.: Laminin 5 deposition regulates keratinocyte polarization and persistent migration. J. Cell Sci. 117(8), 1351–1363 (2004)
Gracheva M., Othmer H.: A continuum model of motility in ameboid cells. Bull. Math. Biol. 66(1), 167–193 (2004)
He X., Dembo M.: On the mechanics of the first cleavage division of the sea urchin egg. Exp. Cell Res. 233(2), 252–273 (1997)
Herant M., Marganski W., Dembo M.: The mechanics of neutrophils: synthetic modeling of three experiments. Biophys. J. 84(5), 3389–3413 (2003)
Hinz B., Alt W., Johnen C., Herzog V., Kaiser H.: Quantifying lamella dynamics of cultured cells by SACED, a new computer-assisted motion analysis. Exp. Cell Res. 251, 234–243 (1999)
Hood P., Taylor C.: A numerical solution of the Navier-Stokes equations using the finite element technique. Comput. Fluids 1, 73–100 (1973)
Hou L., Ravindran S.: A penalized neumann control approach for solving an optimal Dirichlet control problem for the Navier-Stokes equations. SIAM J. Control Optim. 36(5), 1795–1814 (1998)
Jiang G., Giannone G., Critchley D., Fukumoto E.: Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 424, 334–337 (2003)
Jiang G., Peng D.: Weighted ENO schemes for Hamilton–Jacobi equations. SIAM J. Sci. Comput. 21(6), 2126–2143 (1999)
Landau L., Lifshitz E.: Fluid Mechanics, 2nd edn. Butterworth Heinemann, London (1999)
Lauffenburger D., Horwitz A.: Cell migration: a physically integrated molecular process. Cell 84(3), 359–69 (1996)
Lo C., Wang H., Dembo M., Wang Y.: Cell movement is guided by the rigidity of the substrate. Biophys. J. 79(1), 144–152 (2000)
Machacek M., Danuser G.: Morphodynamic profiling of protrusion phenotypes. Biophys. J. 90(4), 1439–1452 (2006)
Mitchison T., Cramer L.: Actin-based cell motility and cell locomotion. Cell 84(3), 371–9 (1996)
Mittal R., Iaccarino G.: Immersed boundary methods. Annu. Rev. Fluid Mech. 37(1), 239–261 (2005)
Mogilner A., Edelstein-Keshet L.: Regulation of actin dynamics in rapidly moving cells: a quantitative analysis. Biophys. J. 83(3), 1237–1258 (2002)
Mogilner A., Oster G.: Cell motility driven by actin polymerization. Biophys. J. 71(6), 3030–3045 (1996)
Mogilner A., Oster G.: Polymer motors: pushing out the front and pulling up the back. Current Biol. 13(18), 721–733 (2003)
Mogilner A., Verzi D.: A simple 1-D physical model for the crawling nematode sperm cell. J. Stat. Phys. 110(3), 1169–1189 (2003)
Möhl, C.: Modellierung von Adhäsions- und Cytoskelett-Dynamik in Lamellipodien migratorischer Zellen. Diploma thesis, Rheinische Friedrich-Wilhelms-Universität Bonn (2005)
Oliver J., King J., McKinlay K., Brown P., Grant D., Scotchford C., Wood J.: Thin-film theories for two-phase reactive flow models of active cell motion. Math. Med. Biol. 22(1), 53 (2005)
Osher S., Shu C.: High-order essentially nonoscillatory schemes for Hamilton-Jacobi equations. SIAM J. Numer. Anal. 28(4), 907–922 (1991)
Palecek S.P., Horwitz A.F., Lauffenburger D.A.: Kinetic model for integrin-mediated adhesion release during cell migration. Ann. Biomed. Eng. 27(2), 219–235 (1999)
Parent C.: A cell’s sense of direction. Science 284(5415), 765–770 (1999)
Parsons J.: Focal adhesion kinase: the first ten years. J. Cell Sci. 116, 1409–1416 (2003)
Plow E., Haas T., Zhang L., Loftus J., Smith J.: Ligand binding to integrins. J. Biol. Chem. 275(29), 21785–21788 (2000)
Pollard T., Borisy G.: Cellular motility driven by assembly and disassembly of actin filaments. Cell 112(4), 453–465 (2003)
Ponti A., Machacek M., Gupton S., Waterman-Storer C., Danuser G.: Two distinct actin networks drive the protrusion of migrating cells. Science 305(5691), 1782–6 (2004)
Rubinstein B., Jacobson K., Mogilner A.: Multiscale two-dimensional modeling of a motile simple-shaped cell. Multiscale Model. Simul. 3, 413 (2005)
Seifert U.: Rupture of multiple parallel molecular bonds under dynamic loading. Phys. Rev. Lett. 84(12), 2750–2753 (2000)
Sethian J.: A fast marching level set method for monotonically advancing fronts. Proc. Natl. Acad. Sci. USA 93(4), 1591–5 (1996)
Sethian J.: Level Set Methods. Cambridge University Press, Cambridge (1996)
Small J., Herzog M., Anderson K.: Actin filament organization in the fish keratocyte lamellipodium. J. Cell Biol. 129(5), 1275–1286 (1995)
Small J., Stradal T., Vignal E., Rottner K.: The lamellipodium: where motility begins. Trends Cell Biol. 12(3), 112–120 (2002)
Thoumine O., Meister J.J.: A probabilistic model for ligand–cytoskeleton transmembrane adhesion: predicting the behavior of microspheres on the surface of migrating cells. J. Theor. Biol. 204(3), 381–392 (2000)
Turner C.: Paxillin and focal adhesion signalling. Nature Cell Biol. 2, E231–E236 (2000)
Verkhovsky A., Svitkina T., Borisy G.: Self-polarization and directional motility of cytoplasm. Curr. Biol. 9(1), 11–20 (1999)
Xian W., Tang J., Janmey P., Braunlin W.: The polyelectrolyte behavior of actin filaments: a 25Mg NMR study. Biochemistry 38(22), 7219–7226 (1999)
Ye T., Mittal R., Udaykumar H., Shyy W.: An accurate cartesian grid method for viscous incompressible flows with complex immersed boundaries. J. Comput. Phys. 156(2), 209–240 (1999)
Zhu C., Skalak R.: A continuum model of protrusion of pseudopod in leukocytes. Biophys. J. 54(6), 1115–1137 (1988)
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
The Below are the Electronic Supplementary Materials.
285_2008_179_MOESM1_ESM.mpg
Movie 1 (related to Fig. 6 of the article): F-actin pattern formation. F-actin pattern formation starting from a constant concentration in a stationary domain. Coloring indicates the volume fraction of F-actin (red: high F-actin concentration, blue: low). Arrows represent the velocity of the F-actin field. ESM 1 (MPG 43,588 kb)
285_2008_179_MOESM2_ESM.mpg
Movie 2 (related to Fig. 7 of the article): Polarization. Polarization of an initially symmetric cell fragment. Coloring indicates the density of actin-and-surface bound integrins (red: high integrin density, blue: low). Arrows represent the velocity of the F-actin field. ESM 2 (MPG 21,385 kb)
Rights and permissions
About this article
Cite this article
Kuusela, E., Alt, W. Continuum model of cell adhesion and migration. J. Math. Biol. 58, 135–161 (2009). https://doi.org/10.1007/s00285-008-0179-x
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-008-0179-x