Abstract
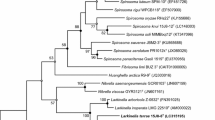
A Gram-negative, short rod-shaped, gliding motile, and pale pink-pigmented bacterial strain, designated 15J9-9T, was isolated from the soil of Iho Tewoo Beach, Jeju Island, Korea, and characterized taxonomically using a polyphasic approach. Comparative 16S rRNA gene sequence analysis showed that strain 15J9-9T belonged to the family Cytophagaceae and was related to Larkinella bovis M2T2B15T (96.5% similarity), Larkinella arboricola Z0532T (95.3% similarity), and Larkinella insperata LMG 22510T (95.2% similarity). The DNA G+C content of strain 15J9-9T was 48 mol%. The detection of phosphatidylethanolamine, phosphatidylserine, two unknown aminophospholipids (APL1 and APL2), and two unknown polar lipids (L1 and L2) in polar lipid profile, menaquinone-7 as the predominant quinone, and a fatty acid profile of C16:1 ω5c, iso-C15:0, and iso-C17:0 3-OH as the major fatty acids supported the affiliation of strain 15J9-9T to the genus Larkinella. Based on its phenotypic properties and phylogenetic distinctiveness, strain 15J9-9T should be classified in the genus Larkinella as a representative of a novel species, for which the name Larkinella harenae sp. nov. is proposed. The type strain is 15J9-9T (=KCTC 42999T = JCM 31656T).

Similar content being viewed by others
References
Vancanneyt M, Nedashkovskaya OI, Snauwaert C, Mortier S, Vandemeulebroecke K, Hoste B, Dawyndt P, Frolova GM, Janssens D, Swings J (2006) Larkinella insperata gen. nov., sp. nov., a bacterium of the phylum ‘Bacteroidetes’ isolated from water of a steam generator. Int J Syst Evol Microbiol 56:237–241
Anandham R, Kwon S, Weon H, Kim S, Kim Y, Gandhi PI, Kim Y, Jee H (2011) Larkinella bovis sp. nov., isolated from fermented bovine products, and emended descriptions of the genus Larkinella and of Larkinella insperata Vancanneyt et al. 2006. Int J Syst Evol Microbiol 61:30–34
Kulichevskaya IS, Zaichikova MV, Detkova EN, Dedysh SN, Zavarzin GA (2009) Larkinella arboricola sp. nov., a new spiral-shaped bacterium of the phylum Bacteroidetes isolated from the microbial community of decomposing wood. Microbiology 78:741–746
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis, version 6.0. Mol Biol Evol 30:2725–2729
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gerhardt P, Murray RGE, Wood WA, Krieg NR (1994) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI, Newark
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Komagata K, Suzuki K (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Collins MD, Jones D (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol Rev 45:316–354
Hiraishi A, Ueda Y, Ishihara J, Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by high performance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–469
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Tamaoka J, Komagata K (1984) Determination of DNA base composition by reversed phase high-performance liquid chromatography. FEMS Microbiol Lett 25:125–128
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Acknowledgements
This work was supported by the Human Resource Training Program for Regional Innovation and Creativity through the Ministry of Education and National Research Foundation (NRF) of Korea (NRF-2014H1C1A1066929) and by the Brain Pool Program of 2016 (Grant 162S-4-3-1727) through the Korean Federation of Science and Technology Societies (KOFST) funded by the Ministry of Science, ICT and Future Planning, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest is declared.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Park, SJ., Lee, JJ., Lee, SY. et al. Larkinella harenae sp. nov., Isolated from Korean Beach Soil. Curr Microbiol 74, 798–802 (2017). https://doi.org/10.1007/s00284-017-1246-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-017-1246-6