Abstract
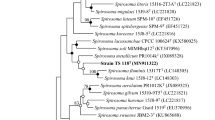
A Gram-negative, short-rod-shaped bacterial strain with gliding motility, designated as DG5AT, was isolated from a rice field soil in South Korea. Phylogenic analysis using 16S rRNA gene sequence of the new isolate showed that strain DG5AT belong to the genus Spirosoma in the family Spirosomaceae, and the highest sequence similarities were 95.5 % with Spirosoma linguale DSM 74T, 93.4 % with Spirosoma rigui WPCB118T, 92.8 % with Spirosoma luteum SPM-10T, 92.7 % with Spirosoma spitsbergense SPM-9T, and 91.9 % with Spirosoma panaciterrae Gsoil 1519T. Strain DG5AT revealed resistance to gamma and UV radiation. Chemotaxonomic data showed that the most abundant fatty acids were summed feature C16:1 ω7c/C16:1 ω6c (36.90 %), C16:1 ω5c (29.55 %), and iso-C15:0 (14.78 %), and the major polar lipid was phosphatidylethanolamine (PE). The DNA G+C content of strain DG5AT was 49.1 mol%. Together, the phenotypic, phylogenetic, and chemotaxonomic data supported that strain DG5AT presents a novel species of the genus Spirosoma, for which the name Spirosoma radiotolerans sp. nov., is proposed. The type strain is DG5AT (=KCTC 32455T = JCM19447T).


Similar content being viewed by others
References
Baik KS, Kim MS, Park SC, Lee DW, Lee SD, Ka JO, Choi SK, Seong CN (2007) Spirosoma rigui sp. nov., isolated from fresh water. Int J Syst Evol Microbiol 57:2870–2873. doi:10.1099/ijs.0.65302-0
Battista JR, Earl AM, Park MJ (1999) Why is Deinococcus radiodurans so resistant to ionizing radiation? Trends Microbiol 7:362–365
Buck JD (1982) Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl Environ Microbiol 44:992–993
Collins MD, Jones D (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev 45(2):316–354
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Filippini M, Kaech A, Ziegler U, Bagheri HC (2011) Fibrisoma limi gen. nov., sp. nov., a filamentous bacterium isolated from tidal flats. Int J Syst Evol Microbiol 61:1418–1424. doi:10.1099/ijs.0.025395-0
Finster KW, Herbert RA, Lomstein BA (2009) Spirosoma spitsbergense sp. nov. and Spirosoma luteum sp. nov., isolated from a high Arctic permafrost soil, and emended description of the genus Spirosoma. Int J Syst Evol Microbiol 59:839–844. doi:10.1099/ijs.0.002725-0
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specified tree topology. Syst Zool 20(4):406–416
Garrity GM, Holt JG (2001) The road map to the manual. In: Garrity GA, Boone DR, Castenholz RW (eds) Bergey’s manual of systematic bacteriology, vol 1, 2nd edn. Springer, New York, pp 119–166
Gosink JJ, Woese CR, Staley JT (1998) Polaribactger gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga–Flavobacterium–Bactgerodes group and reclassification of ‘Flectobacillus glomeraatus’ as Polaribacer glomeratus comb. nov. Int J Syst Bacteriol 48:223–235
Hiraishi A, Ueda Y, Ishihara J, Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by highperformance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–469
Im S, Song D, Joe M, Kim D, Park DH, Lim S (2013) Comparative survival analysis of 12 histidine kinase mutants of Deinococcus radiodurans after exposure to DNA-damaging agents. Bioprocess Biosyst Eng 36:781–789
Jiang F, Xiao M, Chen L, Kan W, Peng F, Dai J, Chang X, Li W, Fang C (2013) Huanghella arctica gen. nov., sp. nov., a bacterium of the family Cytophagaceae isolated from Arctic tundra soil. Int J Syst Evol Microbiol 63:696–702. doi:10.1099/ijs.0.041533-0
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721. doi:10.1099/ijs.0.038075-0
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120
Komagata K, Suzuki KI (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–205
Larkin JM, Borrall R (1978) Spirosomaceae, a new family to contain the Genera Spirosoma migula 1894, Flectobacillus Larkin et al. 1977, and Runella Larkin and Williams 1978. Int J Syst Bacteriol 28(4):595–596
Larkin JM, Borrall R (1984) Deoxyribonucleic acid base compositions, and homology of “microcyclus”, Spirosoma, and similar organisms. Int J Syst Bacteriol 34(2):211–215
Larkin JM, Borrall R (1984) Family 1. Spirosomaceae Larkin and Borrall 1978 595AL. In: Krieg NR, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 1. Williams & Wilkins, Baltimore, pp 125–132
Migula W (1894) Űber ein neues System der Bakterien. Arb Bakt Inst Karlsr 1:235–238
Minnikin DE, O’Donnella AG, Goodfellowb M, Aldersonb G, Athalyeb M, Schaala A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2(5):233–241. doi:10.1016/0167-7012(84)90018-6
Rainey FA, Ray K, Ferreira M, Gatz BZ, Nobre MF, Bagaley D, Rash BA, Park MJ, Earl AM, Shank NC, Small AM, Henk MC, Battista JR, Kampfer P, da Costa MS (2005) Extensive diversity of ionizing-radiation-resistant bacteria recovered from Sonoran Desert soil and description of nine new species of the genus Spirosoma obtained from a single soil sample. Appl Environ Microbiol 71(9):5225–5235. doi:10.1128/AEM.71.9.5225-5235.2005
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc., Newark
Selvam K, Duncan JR, Tanaka M, Battista JR (2013) DdrA, DdrD, and PprA: components of UV and mitomycin C resistance in Deinococcus radiodurans R1. PLoS One 8(7):e69007
Tamaoka J, Komagata K (1984) Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett 25:125–128
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. doi:10.1093/molbev/msr121
Ten LN, Xu JL, Jin FX, Im WT, Oh HM, Lee ST (2009) Spirosoma panaciterrae sp. nov., isolated from soil. Int J Syst Evol Microbiol 59:331–335. doi:10.1099/ijs.0.002436-0
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24):4876–4882. doi:10.1093/nar/25.24.4876
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173(2):697–703
Weon HY, Noh HJ, Son JA, Jang HB, Kim BY, Kwon SW, Stackebrandt E (2008) Rudanella lutea gen. nov., sp. nov., isolated from an air sample in Korea. Int J Syst Evol Microbiol 58:474–478. doi:10.1099/ijs.0.65358-0
Acknowledgments
This work was supported by a special research grant from Seoul Women’s University (2014). The authors are grateful to Dr. Jean Euzéby for help with etymology.
Author information
Authors and Affiliations
Corresponding author
Additional information
The NCBI GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain DG5A (=KCTC 32455 = JCM19447 ) is KF303585.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, JJ., Srinivasan, S., Lim, S. et al. Spirosoma radiotolerans sp. nov., a Gamma-Radiation-Resistant Bacterium Isolated from Gamma Ray-Irradiated Soil. Curr Microbiol 69, 286–291 (2014). https://doi.org/10.1007/s00284-014-0584-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-014-0584-x