Abstract
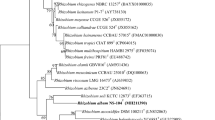
The strain designated as AB21T was isolated from chloroethylenes contaminated soil. Cells are gram-negative, aerobic, non-spore-forming, and motile rods. Phylogenetic analysis based on 16S rRNA gene sequence showed that it belonged to the genus Rhizobium, and was closely related to Rhizobium sullae IS 123T (97.4 %), Rhizobium yanglingense SH 22623T (97.2 %), Rhizobium gallicum R 602spT (97.1 %), Rhizobium alamii GBV 016T (97.0 %), and Rhizobium monogolense USDA 1844T (97.0 %). It showed less than 97 % identity with the remaining Rhizobium species. This novel isolate grew optimally at 25–37 °C (optimum, 30 °C) and pH 6–9 (optimum, pH 8.0). It grew in the presence of 0–4 % (w/v) NaCl, tolerating a 4 % (w/v) NaCl. DNA–DNA hybridization experiment shows less than 53 % binding with closely related Rhizobium. Predominant quinone is ubiquinone (Q-10). The major fatty acids were summed feature 8 (composed of C18:1 ω7c/C18:1 ω6c), C19:0 cyclo ω8c, and C16:0. The G+C molar content is 62.5 mol%. Based on the polyphasic analysis, strain AB21T is referred to be a novel species of the genus Rhizobium for which the name Rhizobium halotolerans sp. nov. is proposed. The type strain is AB21T (=KEMC 224-056T = JCM 17536T).

Similar content being viewed by others
References
Frank B (1989) ÜberdiePilzsymbiosederLeguminosen. Berichte der Deutschen BotanischenGesellschaft 7:332–346
Holt GH, Krieg NR, Sneath PH et al (1994) Bergy’s Manual of Systematic Bacteriology (Ninth edition), pp 78, 95, 112, 120
Young JM, Kuykendall LD, Martinez RE et al (2001) A revision of Rhizobium,with an emended description of the genus,and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R.rhizogenes, R.rubi, R.undicola and R.vitis. Int J Syst Evol Microbiol 51:89–103
Jordan DC (1982) Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow-growing, root nodule bacteria from leguminous plants. Int J Syst Bacteriol 32:136–139
Moran MJ, Zogorski JS, Squillace PJ (2007) Chlorinated solvents in groundwater of the United States. Environ Sci Technol 41:74–81
BCCM™/LMG (2011) Culture Medium 203 Bacteria Catalogue http://bccm.belspo.be/db/media_search_results.php?COLL=LMG&FIELD=NUM&TEXT1=203. Accessed 18 Jan 2012
Doetsch RN (1981) Determinative methods of light microscopy. Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC, pp 21–33
Marmur J (1961) A Procedure of deoxyribonucleic acid from micro-organism. J Mol Biol 3:208–218
Weisberg WG, Barns SM, Pelletier DA et al (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Gaunt MW, Turner SL, Rigottier-Gois L (2001) Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int J Syst Evol Microbiol 51:2037–2048
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Thompson JD, Gibson TJ, Plewniak F et al (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Tamura K, Peterson D, Peterson N et al (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Bio Evol. doi:10.1093/molbev/msr121
Felsenstein J (1985) Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Graham PH, Sadowsky MJ, Keyser HH et al (1991) Proposed minimal standards for the description of new genera and species of root- and stem-nodulating bacteria. Int J syst bacteriol. doi:0020-7713/91/0405582-06
Lin DX, Chen WF, Wang FQ et al (2009) Rhizobium mesosinicum sp. nov., isolated from root nodules of three different legumes. Int J Syst Evol Microbiol 59:1919–1923
Martinez-Romero E, Segovia L, Mercante FM et al (1991) Rhizobium tropici, a novel species modulating Phaseolus vulgar in L.Beans and Leucaena sp. trees. Int J syst Bacteriol. doi:0020-7713/91/030417-10
Freiberg C, Broughton WJ, Perret X et al (1997) Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394–401
Collins MD, Jones D (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol Rev 45:316–354
Minnikin DE, O’Donnell AG, Goodfellow M et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101. MIDI Inc., Newark
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Berge O, Lodhi A, Brandelet G et al (2009) Rhizobium alamii sp. nov., an exopolysaccharide-producing species isolated from legume and non-legume rhizospheres. Int J Syst Evol Microbiol 59:367–372
Heyrman J, Verbeeren J, Peter Schumann P et al (2005) Six novel Arthrobacter species isolated from deteriorated mural paintings. Int J Syst Evol Microbiol 55:1457–1464
Keddie RM, Collins MD, Jones D (1986) Genus Arthrobacter Conn and Dimmick 1947. In: Sneath PH, Mair N, Sharpe, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 2. Williams & Wilkins, Baltimore, pp 1288–1301
Zhang X, Li B, Wang H et al (2011) Rhizobium petrolearium sp.nov., isolated from oil-contaminated soil. Int J Syst Evol Microbiol 62:1871–1876
Acknowledgments
This work was supported by Kyonggi University Research Assistant Fellowship 2013.
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank/EMBL/DDBJ accession number for the 16S rRNA, atpD and recA gene sequences of strain AB21T (=KEMC 224-056T = JCM 17536T) are JX307098, JX878388, and JX872399, respectively.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Diange, E.A., Lee, SS. Rhizobium halotolerans sp. nov., Isolated from Chloroethylenes Contaminated Soil. Curr Microbiol 66, 599–605 (2013). https://doi.org/10.1007/s00284-013-0313-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-013-0313-x