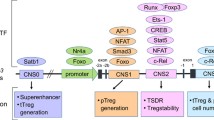
Abstract
The development of T helper cell subsets requires activated T cells that respond to a polarizing cytokine environment resulting in the activation and expression of transcription factors. The subset-specific transcription factors bind and induce the production of specific effector cytokines. Th9 cells express IL-9 and develop in the presence of TGFβ, IL-4, and IL-2. Each of these cytokines activates signaling pathways that are required for Th9 differentiation and IL-9 production. In this review, I summarize what is currently understood about the signaling pathways and transcription factors that promote the Th9 genetic program, providing some perspective for the integration of the signals in regulating the Il9 gene and dictating the expression of other Th9-associated genes. I highlight how experiments in mouse cells have established a transcriptional network that is conserved in human T cells and set the stage toward defining the next important questions for a more detailed understanding of Th9 cell development and function.


Similar content being viewed by others
References
Kaplan MH, Hufford MM, Olson MR (2015) The development and in vivo function of T helper 9 cells. Nat Rev Immunol 15:295–307
Agarwal S, Rao A (1998) Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity 9:765–775
Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH et al (2002) T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol 3:643–651
Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA et al (1998) Helper T cell differentiation is controlled by the cell cycle. Immunity 9:229–237
Murphy KM, Reiner SL (2002) The lineage decisions of helper T cells. Nat Rev Immunol 2:933–944
Grogan JL, Mohrs M, Harmon B, Lacy DA, Sedat JW et al (2001) Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity 14:205–215
Stritesky GL, Yeh N, Kaplan MH (2008) IL-23 mediates stability but not commitment to the Th17 lineage. J Immunol 181:5948–5955
Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ et al (2010) Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature 467:967–971
McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W et al (2007) TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol 8:1390–1397
Tan C, Aziz MK, Lovaas JD, Vistica BP, Shi G et al (2010) Antigen-specific Th9 cells exhibit uniqueness in their kinetics of cytokine production and short retention at the inflammatory site. J Immunol 185:6795–6801
Perumal NB, Kaplan MH (2011) Regulating Il9 transcription in T helper cells. Trends Immunol 32:146–150
Chang HC, Sehra S, Goswami R, Yao W, Yu Q et al (2010) The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol 11:527–534
O'Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y (2011) Genomic views of STAT function in CD4+ T helper cell differentiation. Nat Rev Immunol 11:239–250
O'Shea JJ, Plenge R (2012) JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity 36:542–550
Wei G, Wei L, Zhu J, Zang C, Hu-Li J et al (2009) Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30:155–167
Wei L, Vahedi G, Sun HW, Watford WT, Takatori H et al (2010) Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity 32:840–851
Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V et al (2012) STATs shape the active enhancer landscape of T cell populations. Cell 151:981–993
Pham D, Yu Q, Walline CC, Muthukrishnan R, Blum JS et al (2013) Opposing roles of STAT4 and Dnmt3a in Th1 gene regulation. J Immunol 191:902–911
Yu Q, Thieu VT, Kaplan MH (2007) Stat4 limits DNA methyltransferase recruitment and DNA methylation of the IL-18Ralpha gene during Th1 differentiation. EMBO J 26:2052–2060
Yu Q, Zhou B, Zhang Y, Nguyen ET, Du J et al (2012) DNA methyltransferase 3a limits the expression of interleukin-13 in T helper 2 cells and allergic airway inflammation. Proc Natl Acad Sci U S A 109:541–546
Elo LL, Jarvenpaa H, Tuomela S, Raghav S, Ahlfors H et al (2010) Genome-wide profiling of interleukin-4 and STAT6 transcription factor regulation of human Th2 cell programming. Immunity 32:852–862
Lee DU, Agarwal S, Rao A (2002) Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity 16:649–660
Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W et al (2008) IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(−) effector T cells. Nat Immunol 9:1347–1355
Goswami R, Jabeen R, Yagi R, Pham D, Zhu J et al (2012) STAT6-dependent regulation of Th9 development. J Immunol 188:968–975
Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A et al (2008) Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol 9:1341–1346
Angkasekwinai P, Chang SH, Thapa M, Watarai H, Dong C (2010) Regulation of IL-9 expression by IL-25 signaling. Nat Immunol 11:250–256
Jabeen R, Goswami R, Awe O, Kulkarni A, Nguyen ET et al (2013) Th9 cell development requires a BATF-regulated transcriptional network. J Clin Invest 123:4641–4653
Schmitt E, Germann T, Goedert S, Hoehn P, Huels C et al (1994) IL-9 production of naive CD4+ T cells depends on IL-2, is synergistically enhanced by a combination of TGF-beta and IL-4, and is inhibited by IFN-gamma. J Immunol 153:3989–3996
Gomez-Rodriguez J, Meylan F, Handon R, Hayes ET, Anderson SM et al (2016) Itk is required for Th9 differentiation via TCR-mediated induction of IL-2 and IRF4. Nat Commun 7:10857
Niedbala W, Besnard AG, Nascimento DC, Donate PB, Sonego F et al (2014) Nitric oxide enhances Th9 cell differentiation and airway inflammation. Nat Commun 5:4575
Richard AC, Tan C, Hawley ET, Gomez-Rodriguez J, Goswami R et al (2015) The TNF-family ligand TL1A and its receptor DR3 promote T cell-mediated allergic immunopathology by enhancing differentiation and pathogenicity of IL-9-producing T cells. J Immunol 194:3567–3582
Bassil R, Orent W, Olah M, Kurdi AT, Frangieh M et al (2014) BCL6 controls Th9 cell development by repressing Il9 transcription. J Immunol 193:198–207
Liao W, Spolski R, Li P, Du N, West EE et al (2014) Opposing actions of IL-2 and IL-21 on Th9 differentiation correlate with their differential regulation of BCL6 expression. Proc Natl Acad Sci U S A 111:3508–3513
Yao W, Zhang Y, Jabeen R, Nguyen ET, Wilkes DS et al (2013) Interleukin-9 is required for allergic airway inflammation mediated by the cytokine TSLP. Immunity 38:360–372
Liao W, Schones DE, Oh J, Cui Y, Cui K et al (2008) Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nat Immunol 9:1288–1296
Olson MR, Verdan FF, Hufford MM, Dent AL, Kaplan MH (2016) STAT3 impairs STAT5 activation in the development of IL-9-secreting T cells. J Immunol 196:3297–3304
Murugaiyan G, Beynon V, Pires Da Cunha A, Joller N, Weiner HL (2012) IFN-gamma limits Th9-mediated autoimmune inflammation through dendritic cell modulation of IL-27. J Immunol 189:5277–5283
Vegran F, Berger H, Boidot R, Mignot G, Bruchard M et al (2014) The transcription factor IRF1 dictates the IL-21-dependent anticancer functions of TH9 cells. Nat Immunol 15:758–766
Wang A, Pan D, Lee YH, Martinez GJ, Feng XH et al (2013) Cutting edge: Smad2 and Smad4 regulate TGF-beta-mediated Il9 gene expression via EZH2 displacement. J Immunol 191:4908–4912
Tamiya T, Ichiyama K, Kotani H, Fukaya T, Sekiya T et al (2013) Smad2/3 and IRF4 play a cooperative role in IL-9-producing T cell induction. J Immunol 191:2360–2371
Ebel ME, Kansas GS (2016) Functions of Smad transcription factors in TGF-beta1-induced selectin ligand expression on murine CD4 Th cells. J Immunol 197:2627–2634
Elyaman W, Bassil R, Bradshaw EM, Orent W, Lahoud Y et al (2012) Notch receptors and Smad3 signaling cooperate in the induction of interleukin-9-producing T cells. Immunity 36:623–634
Nakatsukasa H, Zhang D, Maruyama T, Chen H, Cui K et al (2015) The DNA-binding inhibitor Id3 regulates IL-9 production in CD4(+) T cells. Nat Immunol 16:1077–1084
Pham D, Walline CC, Hollister K, Dent AL, Blum JS et al (2013) The transcription factor Twist1 limits T helper 17 and T follicular helper cell development by repressing the gene encoding the interleukin-6 receptor alpha chain. J Biol Chem 288:27423–27433
Wang Y, Bi Y, Chen X, Li C, Li Y et al (2016) Histone deacetylase SIRT1 negatively regulates the differentiation of interleukin-9-producing CD4(+) T cells. Immunity 44:1337–1349
Singh Y, Garden OA, Lang F, Cobb BS (2016) MicroRNAs regulate T-cell production of interleukin-9 and identify hypoxia-inducible factor-2alpha as an important regulator of T helper 9 and regulatory T-cell differentiation. Immunology 149:74–86
Waickman AT, Powell JD (2012) mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol Rev 249:43–58
Goswami R, Kaplan MH (2012) Gcn5 is required for PU.1-dependent interleukin-9 (IL-9) induction in Th9 cells. J Immunol 189:3026–3033
Ramming A, Druzd D, Leipe J, Schulze-Koops H, Skapenko A (2012) Maturation-related histone modifications in the PU.1 promoter regulate Th9-cell development. Blood 119:4665–4674
Gerlach K, Hwang Y, Nikolaev A, Atreya R, Dornhoff H et al (2014) TH9 cells that express the transcription factor PU.1 drive T cell-mediated colitis via IL-9 receptor signaling in intestinal epithelial cells. Nat Immunol 15:676–686
Sehra S, Yao W, Nguyen ET, Glosson-Byers NL, Akhtar N et al (2015) TH9 cells are required for tissue mast cell accumulation during allergic inflammation. J Allergy Clin Immunol 136:433–440
Ahyi AN, Chang HC, Dent AL, Nutt SL, Kaplan MH (2009) IFN regulatory factor 4 regulates the expression of a subset of Th2 cytokines. J Immunol 183:1598–1606
Chang HC, Han L, Jabeen R, Carotta S, Nutt SL et al (2009) PU.1 regulates TCR expression by modulating GATA-3 activity. J Immunol 183:4887–4894
Chang HC, Zhang S, Thieu VT, Slee RB, Bruns HA et al (2005) PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity 22:693–703
Awe O, Hufford MM, Wu H, Pham D, Chang HC et al (2015) PU.1 expression in T follicular helper cells limits CD40L-dependent germinal center B cell development. J Immunol 195:3705–3715
Koh B, Hufford MM, Pham D, Olson MR, Wu T et al (2016) The ETS family transcription factors Etv5 and PU.1 function in parallel to promote Th9 cell development. J Immunol 197:2465–2472
Pham D, Sehra S, Sun X, Kaplan MH (2014) The transcription factor Etv5 controls TH17 cell development and allergic airway inflammation. J Allergy Clin Immunol 134:204–214
Hollenhorst PC, McIntosh LP, Graves BJ (2011) Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem 80:437–471
Staudt V, Bothur E, Klein M, Lingnau K, Reuter S et al (2010) Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity 33:192–202
Ubel C, Sopel N, Graser A, Hildner K, Reinhardt C et al (2014) The activating protein 1 transcription factor basic leucine zipper transcription factor, ATF-like (BATF), regulates lymphocyte- and mast cell-driven immune responses in the setting of allergic asthma. J Allergy Clin Immunol 133(198–206):e191–e199
Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C et al (2007) The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol 8:958–966
Lohoff M, Mittrucker HW, Prechtl S, Bischof S, Sommer F et al (2002) Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc Natl Acad Sci U S A 99:11808–11812
Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B et al (2011) The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol 12:536–543
Schraml BU, Hildner K, Ise W, Lee WL, Smith WA et al (2009) The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 460:405–409
Betz BC, Jordan-Williams KL, Wang C, Kang SG, Liao J et al (2010) Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J Exp Med 207:933–942
Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J et al (2009) Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity 31:941–952
Li P, Spolski R, Liao W, Wang L, Murphy TL et al (2012) BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature 490:543–546
Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W et al (2012) A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science 338:975–980
Ciofani M, Madar A, Galan C, Sellars M, Mace K et al (2012) A validated regulatory network for Th17 cell specification. Cell 151:289–303
Jash A, Sahoo A, Kim GC, Chae CS, Hwang JS et al (2012) Nuclear factor of activated T cells 1 (NFAT1)-induced permissive chromatin modification facilitates nuclear factor-kappaB (NF-kappaB)-mediated interleukin-9 (IL-9) transactivation. J Biol Chem 287:15445–15457
Koch S, Graser A, Mirzakhani H, Zimmermann T, Melichar VO et al (2016) Increased expression of nuclear factor of activated T cells 1 drives IL-9-mediated allergic asthma. J Allergy Clin Immunol 137:1898–1902
Xiao X, Balasubramanian S, Liu W, Chu X, Wang H et al (2012) OX40 signaling favors the induction of T(H)9 cells and airway inflammation. Nat Immunol 13:981–990
Kim IK, Kim BS, Koh CH, Seok JW, Park JS et al (2015) Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells. Nat Med 21:1010–1017
Xiao X, Shi X, Fan Y, Zhang X, Wu M et al (2015) GITR subverts Foxp3(+) Tregs to boost Th9 immunity through regulation of histone acetylation. Nat Commun 6:8266
O'Shea JJ, Paul WE (2010) Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327:1098–1102
Zhou L, Chong MM, Littman DR (2009) Plasticity of CD4+ T cell lineage differentiation. Immunity 30:646–655
Stritesky GL, Muthukrishnan R, Sehra S, Goswami R, Pham D et al (2011) The transcription factor STAT3 is required for Th2 cell development. Immunity 34:39–49
Glosson-Byers NL, Sehra S, Stritesky GL, Yu Q, Awe O et al (2014) Th17 cells demonstrate stable cytokine production in a proallergic environment. J Immunol 193:2631–2640
Xiao X, Shi X, Fan Y, Wu C, Zhang X et al (2016) The costimulatory receptor OX40 inhibits interleukin-17 expression through activation of repressive chromatin remodeling pathways. Immunity 44:1271–1283
Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z et al (2007) Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26:371–381
Lee SH, Chang HS, Jang AS, Park SW, Park JS et al (2011) The association of a single-nucleotide polymorphism of the IL-2 inducible T-cell kinase gene with asthma. Ann Hum Genet 75:359–369
Becker KL, Rosler B, Wang X, Lachmandas E, Kamsteeg M, et al. (2016) Th2 and Th9 responses in patients with chronic mucocutaneous candidiasis and hyper IgE syndrome. Clin Exp Allergy
Yao W, Tepper RS, Kaplan MH (2011) Predisposition to the development of IL-9-secreting T cells in atopic infants. J Allergy Clin Immunol 128:1357–1360
Acknowledgments
This work was supported by Public Health Service grants R01 AI057459 and R03 AI101628 to MHK. Support provided by the Herman B Wells Center was in part from the Riley Children’s Foundation. I thank Drs. R. Nicholas Laribee, Geoffrey Kansas, and Matthew Olson for comments on this review.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is a contribution to the special issue on Th9 Cells in Immunity and Immunopathological Diseases - Guest Editors: Mark Kaplan and Markus Neurath
Rights and permissions
About this article
Cite this article
Kaplan, M.H. The transcription factor network in Th9 cells. Semin Immunopathol 39, 11–20 (2017). https://doi.org/10.1007/s00281-016-0600-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00281-016-0600-2