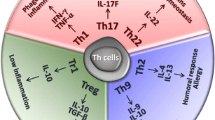
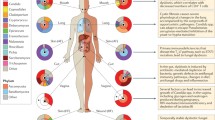
Abstract
An increased understanding of the importance of microbiota in shaping the host’s immune and metabolic activities has rendered fungal interactions with their hosts more complex than previously appreciated. It is now clear that a three-way interaction between host, fungi, and microbiota dictates the types of host–fungus relationship. Indeed, microbial dysbiosis predisposes to a variety of chronic fungal infections and diseases at local and distant sites. By correlating changes in metabolite profiles with microbiota metagenomic composition, we have defined a functional node whereby certain bacteria species contribute to host–fungal symbiosis and mucosal homeostasis. A tryptophan catabolic pathway is exploited by commensal lactobacilli and the mammalian host to increase fitness in response to Candida albicans by inducing resistance and tolerance mechanisms of antifungal immunity. Much like lactobacilli in the gut, Firmicutes change significantly in the airways during aspergillosis. The aryl hydrocarbon receptor has a pivotal role in connecting tryptophan catabolism by microbial communities and the host’s own pathway of tryptophan degradation through the enzyme indoleamine 2,3-dioxygenase 1. These data suggest that the study of the human microbiota in the trans-omics era, with a focus on metagenomics and metabonomics, is providing novel insights into the regulation of host immune responsiveness to fungi.

Similar content being viewed by others
References
Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S (2012) Host-gut microbiota metabolic interactions. Science 336:1262–1267. doi:10.1126/science.1223813
Hooper LV, Littman DR, Macpherson AJ (2012) Interactions between the microbiota and the immune system. Science 336:1268–1273. doi:10.1126/science.1223490
Wang ZK, Yang YS, Stefka AT, Sun G, Peng LH (2014) Review article: fungal microbiota and digestive diseases. Aliment Pharmacol Ther 39:751–766. doi:10.1111/apt.12665
Paulino LC, Tseng CH, Strober BE, Blaser MJ (2006) Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol 44:2933–2941. doi:10.1128/JCM. 00785-06
Cutler JE, Deepe GS Jr, Klein BS (2007) Advances in combating fungal diseases: vaccines on the threshold. Nat Rev Microbiol 5:13–28. doi:10.1038/nrmicro1537
Romani L (2011) Immunity to fungal infections. Nat Rev Immunol 11:275–288. doi:10.1038/nri2939
Underhill DM, Iliev ID (2014) The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol 14:405–416. doi:10.1038/nri3684
Cui L, Morris A, Ghedin E (2013) The human mycobiome in health and disease. Genome Med 5:63. doi:10.1186/gm467
Scanlan PD, Marchesi JR (2008) Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J 2:1183–1193. doi:10.1038/ismej.2008.76
Schulze J, Sonnenborn U (2009) Yeasts in the gut: from commensals to infectious agents. Dtsch Arztebl Int 106:837–842. doi:10.3238/arztebl.2009.0837
Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD (2013) Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One 8:e66019. doi:10.1371/journal.pone.0066019
Chen Y, Chen Z, Guo R, Chen N, Lu H, Huang S, Wang J, Li L (2011) Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis B virus infection. Diagn Microbiol Infect Dis 70:492–498. doi:10.1016/j.diagmicrobio.2010.04.005
Dollive S, Peterfreund GL, Sherrill-Mix S, Bittinger K, Sinha R, Hoffmann C, Nabel CS, Hill DA, Artis D, Bachman MA, Custers-Allen R, Grunberg S, Wu GD, Lewis JD, Bushman FD (2012) A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biol 13:R60. doi:10.1186/gb-2012-13-7-r60
Scupham AJ, Presley LL, Wei B, Bent E, Griffith N, McPherson M, Zhu F, Oluwadara O, Rao N, Braun J, Borneman J (2006) Abundant and diverse fungal microbiota in the murine intestine. Appl Environ Microbiol 72:793–801. doi:10.1128/AEM. 72.1.793-801.2006
Dickson RP, Martinez FJ, Huffnagle GB (2014) The role of the microbiome in exacerbations of chronic lung diseases. Lancet 384:691–702. doi:10.1016/S0140-6736(14)61136-3
Delhaes L, Monchy S, Frealle E, Hubans C, Salleron J, Leroy S, Prevotat A, Wallet F, Wallaert B, Dei-Cas E, Sime-Ngando T, Chabe M, Viscogliosi E (2012) The airway microbiota in cystic fibrosis: a complex fungal and bacterial community—implications for therapeutic management. PLoS One 7:e36313. doi:10.1371/journal.pone.0036313
Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM (2010) Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 6:e1000713. doi:10.1371/journal.ppat.1000713
Ott SJ, Kuhbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, Drews O, Weichert W, Timmis KN, Schreiber S (2008) Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol 43:831–841. doi:10.1080/00365520801935434
Li Q, Wang C, Tang C, He Q, Li N, Li J (2014) Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. J Clin Gastroenterol 48:513–523. doi:10.1097/MCG.0000000000000035
Zhang E, Tanaka T, Tajima M, Tsuboi R, Nishikawa A, Sugita T (2011) Characterization of the skin fungal microbiota in patients with atopic dermatitis and in healthy subjects. Microbiol Immunol 55:625–632. doi:10.1111/j.1348-0421.2011.00364.x
Smeekens SP, Malireddi RK, Plantinga TS, Buffen K, Oosting M, Joosten LA, Kullberg BJ, Perfect JR, Scott WK, van de Veerdonk FL, Xavier RJ, van de Vosse E, Kanneganti TD, Johnson MD, Netea MG (2014) Autophagy is redundant for the host defense against systemic Candida albicans infections. Eur J Clin Microbiol Infect Dis 33:711–722. doi:10.1007/s10096-013-2002-x
Peleg AY, Hogan DA, Mylonakis E (2010) Medically important bacterial-fungal interactions. Nat Rev Microbiol 8:340–349. doi:10.1038/nrmicro2313
Krause R, Schwab E, Bachhiesl D, Daxbock F, Wenisch C, Krejs GJ, Reisinger EC (2001) Role of Candida in antibiotic-associated diarrhea. J Infect Dis 184:1065–1069. doi:10.1086/323550
Erb Downward JR, Falkowski NR, Mason KL, Muraglia R, Huffnagle GB (2013) Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans. Sci Rep 3:2191. doi:10.1038/srep02191
Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y, Tsutsui Y, Qin H, Honda K, Okada T, Hattori M, Fagarasan S (2014) Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 41:152–165. doi:10.1016/j.immuni.2014.05.016
Noverr MC, Huffnagle GB (2004) Does the microbiota regulate immune responses outside the gut? Trends Microbiol 12:562–568
Kalo-Klein A, Witkin SS (1990) Prostaglandin E2 enhances and gamma interferon inhibits germ tube formation in Candida albicans. Infect Immun 58:260–262
Zelante T, Iannitti RG, De Luca A, Arroyo J, Blanco N, Servillo G, Sanglard D, Reichard U, Palmer GE, Latge JP, Puccetti P, Romani L (2012) Sensing of mammalian IL-17A regulates fungal adaptation and virulence. Nat Commun 3:683. doi:10.1038/ncomms1685
Bonifazi P, Zelante T, D’Angelo C, De Luca A, Moretti S, Bozza S, Perruccio K, Iannitti RG, Giovannini G, Volpi C, Fallarino F, Puccetti P, Romani L (2009) Balancing inflammation and tolerance in vivo through dendritic cells by the commensal Candida albicans. Mucosal Immunol 2:362–374. doi:10.1038/mi.2009.17
Romani L, Puccetti P (2006) Protective tolerance to fungi: the role of IL-10 and tryptophan catabolism. Trends Microbiol 14:183–189
Dillon S, Agrawal S, Banerjee K, Letterio J, Denning TL, Oswald-Richter K, Kasprowicz DJ, Kellar K, Pare J, van Dyke T, Ziegler S, Unutmaz D, Pulendran B (2006) Yeast zymosan, a stimulus for TLR2 and dectin-1, induces regulatory antigen-presenting cells and immunological tolerance. J Clin Invest 116:916–928
De Luca A, Montagnoli C, Zelante T, Bonifazi P, Bozza S, Moretti S, D’Angelo C, Vacca C, Boon L, Bistoni F, Puccetti P, Fallarino F, Romani L (2007) Functional yet balanced reactivity to Candida albicans requires TRIF, MyD88, and IDO-dependent inhibition of Rorc. J Immunol 179:5999–6008
Haas-Stapleton EJ, Lu Y, Hong S, Arita M, Favoreto S, Nigam S, Serhan CN, Agabian N (2007) Candida albicans modulates host defense by biosynthesizing the pro-resolving mediator resolvin E1. PLoS One 2:e1316. doi:10.1371/journal.pone.0001316
Gerard R, Sendid B, Colombel JF, Poulain D, Jouault T (2013) An immunological link between Candida albicans colonization and Crohn’s disease. Crit Rev Microbiol. doi:10.3109/1040841X.2013.810587
Standaert-Vitse A, Sendid B, Joossens M, Francois N, Vandewalle-El Khoury P, Branche J, Van Kruiningen H, Jouault T, Rutgeerts P, Gower-Rousseau C, Libersa C, Neut C, Broly F, Chamaillard M, Vermeire S, Poulain D, Colombel JF (2009) Candida albicans colonization and ASCA in familial Crohn’s disease. Am J Gastroenterol 104:1745–1753. doi:10.1038/ajg.2009.225
Noverr MC, Noggle RM, Toews GB, Huffnagle GB (2004) Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun 72:4996–5003. doi:10.1128/IAI. 72.9.4996-5003.2004
Gaitanis G, Velegraki A, Mayser P, Bassukas ID (2013) Skin diseases associated with Malassezia yeasts: facts and controversies. Clin Dermatol 31:455–463. doi:10.1016/j.clindermatol.2013.01.012
Schneider DS, Ayres JS (2008) Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol 8:889–895. doi:10.1038/nri2432
Shapiro H, Thaiss CA, Levy M, Elinav E (2014) The cross talk between microbiota and the immune system: metabolites take center stage. Curr Opin Immunol 30C:54–62. doi:10.1016/j.coi.2014.07.003
Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, Wild B, Camargo SM, Singer D, Richter A, Kuba K, Fukamizu A, Schreiber S, Clevers H, Verrey F, Rosenstiel P, Penninger JM (2012) ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 487:477–481. doi:10.1038/nature11228
Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, Romani L (2013) Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39:372–385. doi:10.1016/j.immuni.2013.08.003
Metz R, Duhadaway JB, Kamasani U, Laury-Kleintop L, Muller AJ, Prendergast GC (2007) Novel tryptophan catabolic enzyme IDO2 is the preferred biochemical target of the antitumor indoleamine 2,3-dioxygenase inhibitory compound D-1-methyl-tryptophan. Cancer Res 67:7082–7087. doi:10.1158/0008-5472.CAN-07-1872
Murray MF (2007) The human indoleamine 2,3-dioxygenase gene and related human genes. Curr Drug Metab 8:197–200
Pfefferkorn ER (1984) Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc Natl Acad Sci U S A 81:908–912
Fallarino F, Grohmann U, Puccetti P (2012) Indoleamine 2,3-dioxygenase: from catalyst to signaling function. Eur J Immunol 42:1932–1937. doi:10.1002/eji.201242572
Munn DH, Mellor AL (2013) Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol 34:137–143. doi:10.1016/j.it.2012.10.001
McGaha TL, Huang L, Lemos H, Metz R, Mautino M, Prendergast GC, Mellor AL (2012) Amino acid catabolism: a pivotal regulator of innate and adaptive immunity. Immunol Rev 249:135–157. doi:10.1111/j.1600-065X.2012.01149.x
Zelante T, Fallarino F, Bistoni F, Puccetti P, Romani L (2009) Indoleamine 2,3-dioxygenase in infection: the paradox of an evasive strategy that benefits the host. Microbes Infect 11:133–141. doi:10.1016/j.micinf.2008.10.007
Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, Sacchettini JC, Behar SM, Rubin EJ (2013) Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 155:1296–1308. doi:10.1016/j.cell.2013.10.045
Grohmann U, Volpi C, Fallarino F, Bozza S, Bianchi R, Vacca C, Orabona C, Belladonna ML, Ayroldi E, Nocentini G, Boon L, Bistoni F, Fioretti MC, Romani L, Riccardi C, Puccetti P (2007) Reverse signaling through GITR ligand enables dexamethasone to activate IDO in allergy. Nat Med 13:579–586. doi:10.1038/nm1563
Yuasa HJ, Ball HJ (2013) Indoleamine 2,3-dioxygenases with very low catalytic activity are well conserved across kingdoms: IDOs of Basidiomycota. Fungal Genet Biol 56:98–106. doi:10.1016/j.fgb.2013.03.003
Bessede A, Gargaro M, Pallotta MT, Matino D, Servillo G, Brunacci C, Bicciato S, Mazza EM, Macchiarulo A, Vacca C, Iannitti R, Tissi L, Volpi C, Belladonna ML, Orabona C, Bianchi R, Lanz TV, Platten M, Della Fazia MA, Piobbico D, Zelante T, Funakoshi H, Nakamura T, Gilot D, Denison MS, Guillemin GJ, DuHadaway JB, Prendergast GC, Metz R, Geffard M, Boon L, Pirro M, Iorio A, Veyret B, Romani L, Grohmann U, Fallarino F, Puccetti P (2014) Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature 511:184–190. doi:10.1038/nature13323
Stockinger B, Di Meglio P, Gialitakis M, Duarte JH (2014) The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol 32:403–432. doi:10.1146/annurev-immunol-032713-120245
Di Meglio P, Duarte JH, Ahlfors H, Owens ND, Li Y, Villanova F, Tosi I, Hirota K, Nestle FO, Mrowietz U, Gilchrist MJ, Stockinger B (2014) Activation of the aryl hydrocarbon receptor dampens the severity of inflammatory skin conditions. Immunity 40:989–1001. doi:10.1016/j.immuni.2014.04.019
Esser C, Bargen I, Weighardt H, Haarmann-Stemmann T, Krutmann J (2013) Functions of the aryl hydrocarbon receptor in the skin. Semin Immunopathol 35:677–691. doi:10.1007/s00281-013-0394-4
Gaitanis G, Magiatis P, Stathopoulou K, Bassukas ID, Alexopoulos EC, Velegraki A, Skaltsounis AL (2008) AhR ligands, malassezin, and indolo[3,2-b]carbazole are selectively produced by Malassezia furfur strains isolated from seborrheic dermatitis. J Investig Dermatol 128:1620–1625. doi:10.1038/sj.jid.5701252
Vlachos C, Schulte BM, Magiatis P, Adema GJ, Gaitanis G (2012) Malassezia-derived indoles activate the aryl hydrocarbon receptor and inhibit toll-like receptor-induced maturation in monocyte-derived dendritic cells. Br J Dermatol 167:496–505. doi:10.1111/j.1365-2133.2012.11014.x
Nguyen LP, Bradfield CA (2008) The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol 21:102–116. doi:10.1021/tx7001965
Fallarino F, Grohmann U, You S, McGrath BC, Cavener DR, Vacca C, Orabona C, Bianchi R, Belladonna ML, Volpi C, Santamaria P, Fioretti MC, Puccetti P (2006) The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J Immunol 176:6752–6761
Lowe MM, Mold JE, Kanwar B, Huang Y, Louie A, Pollastri MP, Wang C, Patel G, Franks DG, Schlezinger J, Sherr DH, Silverstone AE, Hahn ME, McCune JM (2014) Identification of cinnabarinic acid as a novel endogenous aryl hydrocarbon receptor ligand that drives IL-22 production. PLoS One 9:e87877. doi:10.1371/journal.pone.0087877
Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, Bradfield CA (2010) An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol 185:3190–3198. doi:10.4049/jimmunol.0903670
Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller M, Jugold M, Guillemin GJ, Miller CL, Lutz C, Radlwimmer B, Lehmann I, von Deimling A, Wick W, Platten M (2011) An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478:197–203. doi:10.1038/nature10491
Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, Caccamo M, Oukka M, Weiner HL (2008) Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 453:65–71. doi:10.1038/nature06880
Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B (2008) The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453:106–109. doi:10.1038/nature06881
Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD, Colonna M (2012) AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol 13:144–151. doi:10.1038/ni.2187
Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, Zhou L (2012) The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36:92–104. doi:10.1016/j.immuni.2011.11.011
Teunissen MB, Munneke JM, Bernink JH, Spuls PI, Res PC, Te Velde A, Cheuk S, Brouwer MW, Menting SP, Eidsmo L, Spits H, Hazenberg MD, Mjosberg J (2014) Composition of innate lymphoid cell (ILC) subsets in the human skin: enrichment of NCR ILC3 in lesional skin and blood of psoriasis patients. J Investig Dermatol. doi:10.1038/jid.2014.146
Sonnenberg GF, Fouser LA, Artis D (2011) Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol 12:383–390. doi:10.1038/ni.2025
Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, Raffatellu M (2014) The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 40:262–273. doi:10.1016/j.immuni.2014.01.003
Zenewicz LA, Yin X, Wang G, Elinav E, Hao L, Zhao L, Flavell RA (2013) IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol 190:5306–5312. doi:10.4049/jimmunol.1300016
Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, Fu YX, Zhou L (2013) Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity 39:386–399. doi:10.1016/j.immuni.2013.08.002
Zelante T, Iannitti R, De Luca A, Romani L (2011) IL-22 in antifungal immunity. Eur J Immunol 41:270–275. doi:10.1002/eji.201041246
Liu Y, Yang B, Zhou M, Li L, Zhou H, Zhang J, Chen H, Wu C (2009) Memory IL-22-producing CD4+ T cells specific for Candida albicans are present in humans. Eur J Immunol 39:1472–1479. doi:10.1002/eji.200838811
Eyerich K, Eyerich S, Hiller J, Behrendt H, Traidl-Hoffmann C (2010) Chronic mucocutaneous candidiasis, from bench to bedside. Eur J Dermatol 20:260–265. doi:10.1684/ejd.2010.0910
De Luca A, Carvalho A, Cunha C, Iannitti RG, Pitzurra L, Giovannini G, Mencacci A, Bartolommei L, Moretti S, Massi-Benedetti C, Fuchs D, De Bernardis F, Puccetti P, Romani L (2013) IL-22 and IDO1 affect immunity and tolerance to murine and human vaginal candidiasis. PLoS Pathog 9:e1003486. doi:10.1371/journal.ppat.1003486
Fallarino F, Romani L, Puccetti P (In press) AhR: far more than an environmental sensor. Cell Cycle
Dorrestein PC, Mazmanian SK, Knight R (2014) Finding the missing links among metabolites, microbes, and the host. Immunity 40:824–832. doi:10.1016/j.immuni.2014.05.015
Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G (2009) Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 106:3698–3703. doi:10.1073/pnas.0812874106
Heath-Pagliuso S, Rogers WJ, Tullis K, Seidel SD, Cenijn PH, Brouwer A, Denison MS (1998) Activation of the Ah receptor by tryptophan and tryptophan metabolites. Biochemistry 37:11508–11515. doi:10.1021/bi980087p
Lutgendorff F, Akkermans LM, Soderholm JD (2008) The role of microbiota and probiotics in stress-induced gastro-intestinal damage. Curr Mol Med 8:282–298
Tannock GW, Savage DC (1974) Influences of dietary and environmental stress on microbial populations in the murine gastrointestinal tract. Infect Immun 9:591–598
Sjogren YM, Tomicic S, Lundberg A, Bottcher MF, Bjorksten B, Sverremark-Ekstrom E, Jenmalm MC (2009) Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin Exp Allergy 39:1842–1851. doi:10.1111/j.1365-2222.2009.03326.x
Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, Liu C, West ML, Singer NV, Equinda MJ, Gobourne A, Lipuma L, Young LF, Smith OM, Ghosh A, Hanash AM, Goldberg JD, Aoyama K, Blazar BR, Pamer EG, van den Brink MR (2012) Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med 209:903–911. doi:10.1084/jem.20112408
Beck JM, Young VB, Huffnagle GB (2012) The microbiome of the lung. Transl Res 160:258–266. doi:10.1016/j.trsl.2012.02.005
Cui L, Morris A, Huang L, Beck JM, Twigg HL 3rd, von Mutius E, Ghedin E (2014) The microbiome and the lung. Ann Am Thorac Soc 11(Suppl 4):S227–S232. doi:10.1513/AnnalsATS.201402-052PL
Kousha M, Tadi R, Soubani AO (2011) Pulmonary aspergillosis: a clinical review. Eur Respir Rev 20:156–174
Mear JB, Gosset P, Kipnis E, Faure E, Dessein R, Jawhara S, Fradin C, Faure K, Poulain D, Sendid B, Guery B (2014) Candida albicans airway exposure primes the lung innate immune response against Pseudomonas aeruginosa infection through innate lymphoid cell recruitment and interleukin-22-associated mucosal response. Infect Immun 82:306–315. doi:10.1128/IAI. 01085-13
Acknowledgments
This work was supported by the ERC-2011-AdG-293714 to L. Romani. We thank Drs. Cristina Massi Benedetti for the editorial assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is a contribution to the special issue on Immunopathology of Fungal Diseases - Guest Editor: Jean-Paul Latge
Rights and permissions
About this article
Cite this article
Romani, L., Zelante, T., Palmieri, M. et al. The cross-talk between opportunistic fungi and the mammalian host via microbiota’s metabolism. Semin Immunopathol 37, 163–171 (2015). https://doi.org/10.1007/s00281-014-0464-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00281-014-0464-2