Abstract
Terminal disinfection and daily cleaning have been performed in hospitals in Taiwan for many years to reduce the risks of healthcare-associated infections. However, the effectiveness of these cleaning approaches and dynamic changes of surface microbiota upon cleaning remain unclear. Here, we report the surface changes of bacterial communities with terminal disinfection and daily cleaning in a medical intensive care unit (MICU) and only terminal disinfection in a respiratory care center (RCC) using 16s ribosomal RNA (rRNA) metagenomics. A total of 36 samples, including 9 samples per sampling time, from each ward were analysed. The clinical isolates were recorded during the sampling time. A large amount of microbial diversity was detected, and human skin microbiota (HSM) was predominant in both wards. In addition, the colonization rate of the HSM in the MICU was higher than that in the RCC, especially for Moraxellaceae. A higher alpha-diversity (p = 0.005519) and a lower UniFrac distance was shown in the RCC due to the lack of daily cleaning. Moreover, a significantly higher abundance among Acinetobacter sp., Streptococcus sp. and Pseudomonas sp. was shown in the RCC compared to the MICU using the paired t test. We concluded that cleaning changes might contribute to the difference in diversity between two wards.
Similar content being viewed by others
Introduction
Healthcare-associated infections (HAIs) remain a major cause of patient morbidity and mortality (Wang et al. 2007). These infections are mainly caused by several nosocomial pathogens. These include methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus, extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella species, and fluoroquinolone- or carbapenem-resistant Enterobacteriaceae, Pseudomonas and Acinetobacter spp. (Deshpande et al. 2007; Hsueh 2012). In the USA, 1.7 million HAIs resulting in 99,000 deaths were reported in 2002 (Klevens et al. 2007). Inanimate hospital environments (IHEs) are often overlooked reservoirs for these bacteria. Transmission of healthcare-associated pathogens (HAPs) via surface contamination is now a critical issue in HAIs (Bokulich et al. 2013, Strassle et al. 2012, Weber et al. 2010). Thus, environmental cleaning and strict monitoring of IHEs is necessary to drastically reduce the risk of HAIs.
In recent years, a number of studies have demonstrated that environmental cleaning interventions can improve the effectiveness of cleaning and reduce contamination on surfaces (Dumigan et al. 2010; Eckstein et al. 2007). Infections, including urinary tract infections, bloodstream infections and ventilator-associated pneumonia, constitute one of the major issues in intensive care units (ICUs) and respiratory care centers (RCCs) (Kuo et al. 2008; Liu et al. 2011). Patients in ICUs are more susceptible to colonization, and the organisms are often more resistant to antimicrobial agents than in other environments. Furthermore, stabilized patients in ICUs will be transferred to RCCs if they require ventilator use for more than 21 days in the ICU. The Centres for Disease Control and Prevention (CDC) infection control guidelines for the routine and terminal cleaning of hospital rooms recommend paying attention to the disinfection of patient care surfaces, especially surfaces designated as ‘high-touch’, as a source of HAIs (Sehulster and Chinn 2003). Daily cleaning is conducted once each day, and terminal cleaning is conducted only when a patient is discharged from the room in clinical practice. In Taiwan, terminal disinfection is required for all of the wards in hospitals; however, whether daily cleaning is performed or not depends on the infection control policy of the individual hospital. Although the effectiveness of cleaning on repeatedly touched hospital surfaces has been demonstrated in several countries (Donskey 2013), the higher incidence of HAPs, e.g. Acinetobacter baumannii, causing higher mortality and morbidity was previously reported in Taiwan (Chen et al. 2015), and the prevalence of carbapenem-resistant A. baumannii (CRAB) has been gradually rising in Taiwan (Jean and Hsueh 2011b; Jean et al. 2009), China (He et al. 2011) and Korea (Lee et al. 2004). Thus, evaluation of the effectiveness of environmental cleaning upon daily cleaning or after terminal disinfection in hospitals would be required in Taiwan to reduce the incidence of HAIs.
The effectiveness of cleaning on IHEs was routinely evaluated by standard cultivation techniques. However, more than 99 % of the microorganisms present in many natural environments are not readily cultivable (Amann et al. 1995). Recently, culture-independent methods based on amplification and sequencing of bacterial metagenomes have been developed to identify thousands of different bacteria in a single sample (Tringe and Hugenholtz 2008). Using 16s ribosomal RNA (rRNA) amplicon metagenomics, the identification and tracking of bacterial diversity in hospital environments has become feasible (Bokulich et al. 2013; Hewitt et al. 2013; Lo et al. 2011; Oberauner et al. 2013).
Here, we used 16s rRNA amplicon sequencing and quantitative PCR (qPCR) to investigate the microbial diversity of IHEs in a medical intensive care unit (MICU) and an RCC in a study hospital. Two cleaning procedures were performed in the two wards; the MICU had daily cleaning and terminal disinfection, and the RCC had terminal disinfection. The purpose of this study was to (1) investigate the bacterial community diversity with two different cleaning methods in the MICU and RCC during the study period and (2) to elucidate the relationship of persistent A. baumannii colonization under two different hospital cleaning methods in two wards.
Materials and methods
Patients and setting
Hsin-Chu Branch of National Taiwan University Hospital (NTUH) is a regional teaching hospital with 699 ward beds in northern Taiwan. There were 16 beds in MICU and 12 beds in the RCC. Clinical data were collected during the sampling period in February 2014 in both wards of NTUH Hsin-Chu Branch.
Disinfection procedure
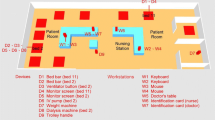
Both MICU and RCC underwent terminal disinfection when a patient was discharged or changed his bed. The terminal disinfection procedure was performed as follows: (i) a health care workers (HCWs) wiped the surface (the bed, monitor, ventilator, and stethoscope) with 500-ppm hypochlorite and kept it wet for 30 min and then wiped it again with clean water; (ii) a HCWs changed the curtain, mattress, and quilt. In addition, daily cleaning was performed everyday in each bed in MICU, which include wiping the bed bar monitor, ventilator, stethoscope, oxygen supply and suction button, hemodialysis machine, intravenous pump and feeding pump with the 500 ppm hypochlorite only without waiting 30 min to wipe with water. The only surfaces not cleaned with the anti-bacterial wipes were the keyboards and mice of the computers. Table 1 and Fig. 1 list the location of the surfaces sampled and the disinfection procedure in the two wards.
Sample collection and DNA extraction
Environmental samples were collected from the MICU and RCC at the Hsin-Chu Branch of NTUH. A list of all of the tested sites is provided in Fig. 1. Sampling sites around a bed in each ward were chosen based on the frequency with which the surfaces were highly touched. The sampling times were performed as a patient was hospitalized in the MICU or RCC at 0, 3, 7, and 14 days. The sampling time for day 0 is designated as around 30 min for sampling after terminal disinfection and before a new patient was admitted to the ward. The sampling time for days 3, 7, and 14 was around 20 h after daily cleaning in the MICU. The surfaces were sampled with sterile cotton swabs. On the flat surfaces (e.g., ventilator screen and monitor screen), approximately 12 cm2 of each surface was swabbed. The computer mice were swabbed in their entirety, and a total of 10 keys on each keyboard were swabbed. After sampling, the swabs were transported back to the microbiology laboratory at Yuanpei University for further DNA extraction.
DNA extraction and PCR
The protocols for DNA extraction and PCR were followed based on the description by Tang et al. (2015). Briefly, and prior to DNA extraction, the cotton from the swab was removed, the DNA was extracted and barcoded PCR amplification was performed using the V1 forward primer (5′-AGAGTTTGATCCTGGCTCAG-3′) and the V2 reverse primers (5′-TGCTGCCTCCCGTAGGAGT-3′) with 382-bp amplicons flanking the highly variable V1–V2 region of the 16s rRNA gene sequence (Cai et al. 2013).
Amplicon sequencing
Individual barcoded PCR products were purified and then pooled with a total combined concentration of 1 μg (total volume = 50 μl). All of the barcoded PCR fragments were sequenced using an Illumina Miseq Desktop Sequencer at Tri-I Biotech Inc. (Taipei, Taiwan). The raw sequences were deposited at the NCBI sequence Read Archive under the Bioproject accession number PRJNA301917.
Quantitative PCR
The amount of net bacterial biomass in a sample was determined by qPCR using an Eco Real-Time PCR System (Illumina, CA, USA). A pair of primers, V1 and V2, for the 16s rRNA gene were used. Briefly, the template DNA extracted from the samples was diluted 1:10, and 2.5 μl was added to SYBR Green PCR Master Mix (Biogenesis Technologies, Inc., Taiwan) for each reaction that was used for analysis. A melting curve analysis was performed to determine the specificity of the PCR products. Genomic DNA from E. coli ATCC 25922 was used to generate the standard curves for quantification. All of the samples were run in triplicate to confirm the reproducibility of the data.
Computational and statistical analyses
Paired-end reads were assembled with PEAR software (http://www.exelixis-lab.org/web/software/pear), and low-quality reads were filtered using the QIIME database with default parameters (Caporaso et al. 2010). The remaining reads were checked for chimeric sequences with UCHIME (Edgar et al. 2011) and clustered into operational taxonomic units (OTUs) using a closed-reference OTU selection protocol at the 97 % identity level with a UPARSE algorithm (Edgar 2013) run against the Greengenes database (McDonald et al. 2012). The taxonomy associated with each OTU was assigned as the taxonomy associated with the reference sequence defining the OTU.
Statistical analysis
All of the statistical analyses were performed using the R project version 3.1.2 (http://www.R-project.org). The numbers of reads for each microbe whose relative abundance was in the top 33 abundant families was represented by bar charts. The bacterial diversity was determined using the Shannon diversity index box plot. One-tailed paired-sample t tests were used to test whether the abundance (sequence reads) of known HAPs differed significantly at days 0 and 14 of hospitalization in both wards. The unweighted UniFrac metric (Lozupone and Knight 2005), which only accounts for the presence/absence of taxa and not abundance, was used to determine the phylogenetic similarity of the bacterial communities associated with the various sampling surfaces. Principal coordinate analysis (PCoA) is used to reduce the dimension of variables performed at the genus level to summarize similarity with different disinfection procedures (Huang and Hsueh 2008). A heatmap was generated based on the description of Ling (1973).
Results
Sample collection and sequencing
A total of 36 samples for each ward, including 9 samples per sampling time, were collected, followed by DNA extraction and barcoded PCR amplification; a high number of amplicons were obtained and sequenced. In total, the raw dataset of 72 samples contained 2,452,954 sequences. Approximately 11,479 sequences per sample were obtained from the RCC, with an average of 328 phylotypes per sample. A total of 13,970 sequences per sample were obtained from the MICU, with an average of 347 phylotypes per sample (Table S1). A phylotype is defined here as organisms sharing ≧97 % 16s rRNA gene sequence identity.
Taxonomic composition of the hospital environmental microbiota and information of clinical isolates
The clinical isolates recorded during the study period in the two wards are listed in Table 2. A total of seven and three isolates was recorded in the MICU and RCC, respectively. A. baumannii, Pseudomonas spp. and Staphylococcus spp. were isolated in both wards during the sampling period.
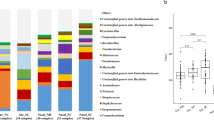
Figure 2 shows the relative abundances of the bacterial communities at the family level in the MICU and the RCC. The abundances of bacterial communities from all of the samples collected for each of the wards was compared (Fig. 2a). Human skin microbiota (HSM) was predominant in both wards compared to the hospital environmental microbiota (HEM). In addition, the colonization rate of HSM in the MICU was higher than that in the RCC, especially for Moraxellaceae (1.0–54.6 %), Corynebacteriaceae (1.1–38.4 %), Prevotellaceae (1.6–41.0 %), Propionibacteriaceae (0.8–46.5 %), Clostridiaceae (1.6–50.2 %), Veillonellaceae (4.5–23.2 %), and Streptococcaceae (2.5–42.8 %). Notably, more than 20 % of Moraxellaceae was observed in the MICU even though the wards were cleaned daily. Figure 2b compares the bacterial profiles between days 0 and 14 of hospitalization in the two wards. Although the two wards had terminal disinfection at day 0, the HSM, such as Corynebacteriaceae and Moraxellaceae, remained the most abundant in these two areas. The HEM, such as Bacillaceae (0.1–55.8 %) and Weeksellaceae (0.3–64.7 %), were also abundant in the RCC. A rapid increase of Porphyromonadaceae was found in both wards at day 14. Furthermore, the association between clinical isolates and environmental communities showed that Moraxellaceae was predominant across the MICU and the RCC at day 14.
Relative abundances of the most abundant bacterial groups on surfaces in the MICU and RCC. Mean values of the samples collected from days 0 and day in the two wards. Their relative height represents the percentage of the reads that can be placed at a family level of taxonomy using Miseq reads with a BLASTX search of the Greengenes database
Dynamic changes between hospital environmental microbiota under two different hospital environmental cleaning methods
We investigated whether two cleaning protocols were related to microbial diversity. An inter- and an intra-ward comparison of the bacterial communities revealed no difference in alpha-diversity (Fig. 3). However, communities from the RCC showed higher alpha-diversity than those in the MICU on day 14 (p = 0.005519), which implied that the lack of daily cleaning in the RCC would contribute to the richness of the bacterial communities.
To further explore the relationship for all of the samples between different bacterial communities in the RCC and the MICU, PCoA analysis was performed using genus-level taxonomic profiles. As shown in Fig. 4, the visual pattern between the MICU and the RCC was quite different. Bacterial communities in the RCC were localized in the positive direction of principal component 2 (PC2) relative to those of the MICU. Notably, four samples taken from the ventilator screen, including M-2-0, M-2-3, M-2-7 and M-2-14, were separated from each other (Fig. 4a). Microbial communities and net bacterial biomass showed that Moraxellaceae became the predominant bacteria after day 3 and was possibly cross-transmitted from the neighbourhood area. Furthermore, net bacterial biomass from non-disinfection areas such as keyboards at both wards were significantly higher (P < 0.05) than the disinfection areas such as ventilator screens (Fig. S1).
PCoA analysis of MICU and RCC samples. a The two principle coordinates explained approximately 43 and 35 % of the variation in the samples from the MICU and RCC, respectively. Principal coordinate analysis of bacterial communities at the genus level from the sampling sites: terminal disinfection and daily cleaning (green), terminal disinfection (blue) or no disinfection (red). b Relative abundances of bacterial communities and net bacterial biomass in the ventilation screen (M-2 site) in the MICU. The net bacterial biomass was determined by qPCR. The orange arrow bar represents the distribution of M-2 samples in the MICU. M-2-1, M-2-3, M-2-7 and M-2-14 represent sampling of the M-2 site at days 0, 3, 7 and 14
A pairwise investigation of the UniFrac distance of bacteria communities associated with various surfaces in the two wards on day 14 of hospitalization is shown in Table 3. The diversity distance is higher in the MICU than in the RCC, implying that the MICU was highly diverse in species on day 14. In total, a higher difference in bacterial composition was observed in the MICU (31/36, 86.1 %) compared to the RCC (17/36, 47.2 %) as the values of the UniFrac distance were more than 0.5.
Occurrence of predominant Acinetobacter species between the hospital environmental microbiota profile and clinical isolates
Because Acinetobacter spp. remains the predominant genus in the two wards, the correlation between Acinetobacter and bacterial communities in IHEs and their association with clinical isolates across 36 samples in each ward is shown in Fig. 5. In the MICU, Acinetobacter spp. and Streptococcus spp. were clustered apart from Staphylococcus, Pseudomonas and Enterococcus. In contrast, Acinetobacter spp. and Staphylococcus spp. was clustered apart from Pseudomonas spp. in the RCC. This result implies that the abundance and appearance of Acinetobacter spp. and their clustered genera differed between the two wards.
Comparison of five different healthcare associated pathogens under two different hospital environmental cleaning methods
The results of the paired t test examining the bacterial abundance of taxa in the MICU and RCC are listed at Table 4. Comparing five different HAPs in the same ward at the two time-points (days 0 and 14), a significantly higher abundance among Acinetobacter spp., Streptococcus spp. and Pseudomonas spp. was shown in the RCC compared to the MICU. Considering two time-points at the different wards, Acinetobacter spp. was significantly abundant in day 0 in the MICU compared to the RCC. However, Staphylococcus spp. and Enterococcus spp. became more abundant in the RCC on day 14 compared to the MICU.
Discussion
Pathogens with a high incidence of antimicrobial resistance in hospitals have become a public health concern (Hsueh et al. 2001). Due to the limited effectiveness of antimicrobials against these pathogens, infection control plays a major role in reducing HAIs. Previously, we applied 16s rRNA metagenomics to evaluate the surface microbiota in the workstation and device within the RCC in this study hospital (Tang et al. 2015). We concluded that bacteria communities in the two areas were clustered together, which indicate that co-transmission via HCWs across workplaces and devices may occur. In this study, we further describe and compare the bacterial community with different hospital cleaning methods in two wards by analysing the dynamics of the surface microbiota in IHEs in this hospital. This is the first study in Taiwan to apply 16s rRNA metagenomics to evaluate the effectiveness of cleaning approaches. In addition, we also elucidate the causality of persistent A. baumannii colonization.
In the present study, we established that (1) hospital environment microbiota can become durably contaminated after exposure to colonized patients at two wards; and (2) although human skin microbiota may become complex within an institution, Acinetobacter spp. are persistently the predominant bacteria in the IHEs in this hospital. The prevalence of CRAB has been gradually rising in Asia, including Taiwan (Jean and Hsueh 2011b; Jean et al. 2009), China (He et al. 2011) and Korea (Lee et al. 2004). Oxacillinase genes, which are located on plasmids and are downstream of insertion sequence ISAba3 and IS1008 (two promoters for transcriptional control of bla OXA genes), were shown to play an important role in Taiwan CRAB isolates (Lee et al. 2009). Because of the serious problems of clonal dissemination of clinical CRABs in Asian countries (Jean et al. 2009; Yang et al. 2009), the incidence of CRAB isolates is highly persistent in Taiwan (Jean and Hsueh 2011a), especially in this study hospital (Liu et al. 2013). Weber’s report showed that Acinetobacter spp. survive for prolonged periods of time in the environment (Weber et al. 2010), and our previous report showed that environmental Acinetobacter spp. remain susceptible to carbapenem even though this microorganism was predominant in this hospital (Tang et al. 2015). Despite the fact that carbapenem-susceptible A. baumannii is widely distributed in these hospitals, these bacteria remain a risk factor for infection due to their natural competence (Ramirez et al. 2010) and their genome plasticity (Mugnier et al. 2009) upon exposure to antibiotics.
There is high variation and a trend of higher richness in the RCC because of a lack of daily cleaning and gradual accumulation of hospital environmental bacteria. Acinetobacter spp. and Streptococcus spp. became the same clonal abundant cluster in the MICU. Under two different cleaning methods, we thought that this association could also reflect an important underlying ecological relationship in the two wards, and the reason is as follows: (1) the accumulation of bacterial communities in the MICU, which had daily cleaning and terminal disinfection, showed a model of ecological succession and Acinetobacter spp. replacing other successional species to become the abundant bacteria in MICU; and (2) the accumulation of bacterial abundance in the RCC, which had terminal disinfection only, showed a different model of ecological succession. In this regard, further experiments are necessary to elucidate these differences.
There were statistically significant differences in bacterial abundance among Enterococcus spp., Acinetobacter spp., Streptococcus spp., and Pseudomonas spp. under two different cleaning methods and between the two time-points in the two wards. In regards to terminal disinfection, effective terminal disinfection significantly decreased environmental contamination of A. baumannii in Strassle’s study (Strassle et al. 2012); however, persistent contamination represents a significant risk factor for transmission of HAIs. Concerning daily cleaning, Hess’s report showed that daily cleaning of ICU rooms, which were occupied by patients colonized with multi-drug resistant A. baumannii (MDRAB), was associated with a non-significant reduction in contamination of healthcare worker gowns and gloves after routine patient care activities (Hess et al. 2013). As shown in Table 4, we assumed that daily environmental cleaning combined with terminal disinfection would lead to significant reductions of healthcare-associated pathogens and fewer MDRAB infections. In Taiwan, terminal disinfection is recommended in all of the wards, but daily cleaning is not necessary for all of the hospitals. For example, daily cleaning was performed in ICUs and RCCs in most of the medical centers and was then gradually decreased in regional hospitals. Further research is needed to determine this policy.
Nevertheless, there are also some limitations to this study (mostly because of the few number of cases). First, only two study wards at a hospital were included. More sample sites from other hospitals would strengthen the general applicability of our results. Second, there is lack of clone identity evidence to confirm the clonal relationship between isolations from hospital environment surfaces and patients; hence, an assessment of causality among clinical isolates and HEM was not able to be analysed. Lastly, there were several variables, including the type of care patients and the different healthcare workers with different compliance between those two wards, which could interfere with the study results. Further studies should be performed to define the role of HEM and HSM and a long-term investigation is necessary to manipulate the cleaning regimens in order to improve the effectiveness of cleaning on hospital environment.
Our study discovered unexpectedly high bacterial community diversity, and hospital environmental microbiota are closely associated with the HSM in IHEs in the MICU and RCC. Although most hospital environmental microbiota can be considered non-pathogenic under normal circumstances, there are potential risks in MICU and RCC settings where patients are extremely vulnerable to infections. Our study also suggests that cleaning changes might contribute to the difference in diversity between two wards. In Taiwan, strict compliance with hospital environmental cleaning policy and the close and continuous monitoring of HAPs at healthcare institutions would be required to reduce HAIs.
References
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59(1):143–169
Bokulich NA, Mills DA, Underwood MA (2013) Surface microbes in the neonatal intensive care unit: changes with routine cleaning and over time. J Clin Microbiol 51(8):2617–2624
Cai L, Ye L, Tong AH, Lok S, Zhang T (2013) Biased diversity metrics revealed by bacterial 16S pyrotags derived from different primer sets. PLoS One 8(1):e53649
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336
Chen CH, Lin LC, Chang YJ, Liu CE, Soon MS (2015) Long-term effectiveness of infection and antibiotic control programs on the transmission of carbapenem-resistant Acinetobacter calcoaceticus-Acinetobacter baumannii complex in Central Taiwan. Med Mal Infect 45(7):264–272
Deshpande LM, Fritsche TR, Moet GJ, Biedenbach DJ, Jones RN (2007) Antimicrobial resistance and molecular epidemiology of vancomycin-resistant Enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn Microbiol Infect Dis 58(2):163–170
Donskey CJ (2013) Does improving surface cleaning and disinfection reduce health care-associated infections? Am J Infect Control 41(5 Suppl):S12–S19
Dumigan DG, Boyce JM, Havill NL, Golebiewski M, Balogun O, Rizvani R (2010) Who is really caring for your environment of care? Developing standardized cleaning procedures and effective monitoring techniques. Am J Infect Control 38(5):387–392
Eckstein BC, Adams DA, Eckstein EC, Rao A, Sethi AK, Yadavalli GK, Donskey CJ (2007) Reduction of Clostridium difficile and vancomycin-resistant Enterococcus contamination of environmental surfaces after an intervention to improve cleaning methods. BMC Infect Dis 7:61
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10(10):996–998
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200
He C, Xie Y, Fan H, Kang M, Tao C, Zhang R, Hu Y, Chen Z, Wang L (2011) Spread of imipenem-resistant Acinetobacter baumannii of European clone II in western China. Int J Antimicrob Agents 38(3):257–260
Hess AS, Shardell M, Johnson JK, Thom KA, Roghmann MC, Netzer G, Amr S, Morgan DJ, Harris AD (2013) A randomized controlled trial of enhanced cleaning to reduce contamination of healthcare worker gowns and gloves with multidrug-resistant bacteria. Infect Control Hosp Epidemiol 34(5):487–493
Hewitt KM, Mannino FL, Gonzalez A, Chase JH, Caporaso JG, Knight R, Kelley ST (2013) Bacterial diversity in two neonatal intensive care units (NICUs. PLoS One 8(1):e54703
Hsueh PR (2012) Study for monitoring antimicrobial resistance trends (SMART) in the Asia-Pacific region, 2002-2010. Int J Antimicrob Agents 40(Suppl):S1–S3
Hsueh PR, Liu YC, Yang D, Yan JJ, Wu TL, Huang WK, Wu JJ, Ko WC, Leu HS, Yu CR, Luh KT (2001) Multicenter surveillance of antimicrobial resistance of major bacterial pathogens in intensive care units in 2000 in Taiwan. Microb Drug Resist 7(4):373–382
Huang YT, Hsueh PR (2008) Antimicrobial drug resistance in Taiwan. Int J Antimicrob Agents 32(Suppl 3):S174–S178
Jean SS, Hsueh PR (2011a) Antimicrobial drug resistance in Taiwan. J Formos Med Assoc 110(1):4–13
Jean SS, Hsueh PR (2011b) High burden of antimicrobial resistance in Asia. Int J Antimicrob Agents 37(4):291–295
Jean SS, Hsueh PR, Lee WS, Chang HT, Chou MY, Chen IS, Wang JH, Lin CF, Shyr JM, Ko WC, Wu JJ, Liu YC, Huang WK, Teng LJ, Liu CY (2009) Nationwide surveillance of antimicrobial resistance among non-fermentative gram-negative bacteria in intensive care units in Taiwan: SMART programme data 2005. Int J Antimicrob Agents 33(3):266–271
Klevens RM, Edwards JR, Richards CL Jr, Horan TC, Gaynes RP, Pollock DA, Cardo DM (2007) Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep 122(2):160–166
Kuo LC, Yu CJ, Kuo ML, Chen WN, Chang CK, Lin HI, Chen CC, Lu MC, Lin CH, Hsieh WF, Chen LW, Chou Y, Huang MS, Lee CH, Chen SC, Thai SL, Chen PC, Chen CH, Tseng CC, Chen YS, Hsiue TR, Hsueh PR (2008) Antimicrobial resistance of bacterial isolates from respiratory care wards in Taiwan: a horizontal surveillance study. Int J Antimicrob Agents 31(5):420–426
Lee SO, Kim NJ, Choi SH, Hyong Kim T, Chung JW, Woo JH, Ryu J, Kim YS (2004) Risk factors for acquisition of imipenem-resistant Acinetobacter baumannii: a case-control study. Antimicrob Agents Chemother 48(1):224–228
Lee YT, Huang LY, Chen TL, Siu LK, Fung CP, Cho WL, Yu KW, Liu CY (2009) Gene cassette arrays, antibiotic susceptibilities, and clinical characteristics of Acinetobacter baumannii bacteremic strains harboring class 1 integrons. J Microbiol Immunol Infect 42(3):210–219
Ling rL (1973) A computer generated aid for cluster analysis. Commun ACM 16(6):355
Liu KS, Wang YT, Lai YC, Yu SF, Huang SJ, Huang HJ, Lu MC, Hsueh PR (2011) Antimicrobial resistance of bacterial isolates from respiratory care wards in Taiwan: a horizontal surveillance study comparison of the characteristics of nosocomial infection and antimicrobial-resistant bacteria in adult intensive care units and two respiratory care facilities for mechanically ventilated patients at a tertiary care centre in Taiwan. Int J Antimicrob Agents 37(1):10–15
Liu CC, Tang CY, Kuo HY, CW L, Chang KC, Liou ML (2013) The origin of Acinetobacter baumannii TYTH-1: a comparative genomics study. Int J Antimicrob Agents 41(4):318–324
Lo WT, Lin WJ, Chiueh TS, Lee SY, Wang CC, Lu JJ (2011) Changing trends in antimicrobial resistance of major bacterial pathogens, 1985-2005: a study from a medical center in northern Taiwan. J Microbiol Immunol Infect 44(2):131–138
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71(12):8228–8235
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P (2012) An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6(3):610–618
Mugnier PD, Poirel L, Nordmann P (2009) Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J Bacteriol 191(7):2414–2418
Oberauner L, Zachow C, Lackner S, Hogenauer C, Smolle KH, Berg G (2013) The ignored diversity: complex bacterial communities in intensive care units revealed by 16S pyrosequencing. Sci Rep 3:1413
Ramirez MS, Don M, Merkier AK, Bistue AJ, Zorreguieta A, Centron D, Tolmasky ME (2010) Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J Clin Microbiol 48(4):1488–1490
Sehulster L, Chinn RY (2003) Guidelines for environmental infection control in health-care facilities. Recommendations of CDC and the healthcare infection control practices advisory committee (HICPAC. MMWR Recomm Rep 52(RR-10):1–42
Strassle P, Thom KA, Johnson JK, Leekha S, Lissauer M, Zhu J, Harris AD (2012) The effect of terminal cleaning on environmental contamination rates of multidrug-resistant Acinetobacter baumannii. Am J Infect Control 40(10):1005–1007
Tang CY, Yiu SM, Kuo HY, Tan TS, Liao KH, Liu CC, Hon WK, Liou ML (2015) Application of 16S rRNA metagenomics to analyze bacterial communities at a respiratory care centre in Taiwan. Appl Microbiol Biotechnol 99(6):2871–2881
Tringe SG, Hugenholtz P (2008) A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol 11(5):442–446
Wang H, Guo P, Sun H, Yang Q, Chen M, Xu Y, Zhu Y (2007) Molecular epidemiology of clinical isolates of carbapenem-resistant Acinetobacter spp. from Chinese hospitals. Antimicrob Agents Chemother 51(11):4022–4028
Weber DJ, Rutala WA, Miller MB, Huslage K, Sickbert-Bennett E (2010) Role of hospital surfaces in the transmission of emerging health care-associated pathogens: norovirus, Clostridium difficile, and Acinetobacter species. Am J Infect Control 38(5 Suppl 1):S25–S33
Yang HY, Lee HJ, Suh JT, Lee KM (2009) Outbreaks of imipenem resistant Acinetobacter baumannii producing OXA-23 beta-lactamase in a tertiary care hospital in Korea. Yonsei Med J 50(6):764–770
Acknowledgment
We would like to thank Mr. Kuan-Hsueh Chen, Mr. Ki-Hok Liao for their technical assistance. We thank Prof. Chuan Yi Tang for his financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
The present work was partially supported by a grant from the Ministry of Science and Technology (grant MOST 104-2627-M-126-001 and grant MOST-104-2627-M-009-006) and Ministry of Health and Welfare Hospital Research Project (104-01).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This study was approved by the Ethics Committee of the NTUH Hsin-Chu Branch (NTUH IRB No. 103-014-E).
Additional information
Chang-Hua Chen and Chi-Chao Tu contributed equally in this work.
Electronic supplementary material
ESM 1
(PDF 198 kb)
Rights and permissions
About this article
Cite this article
Chen, CH., Tu, CC., Kuo, HY. et al. Dynamic change of surface microbiota with different environmental cleaning methods between two wards in a hospital. Appl Microbiol Biotechnol 101, 771–781 (2017). https://doi.org/10.1007/s00253-016-7846-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7846-4