Abstract
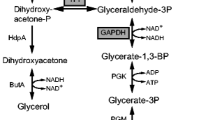
Rapid sugar consumption is important for the microbial production of chemicals and fuels. Here, we show that overexpression of the NADH dehydrogenase gene (ndh) increased glucose consumption rate in Corynebacterium glutamicum under oxygen-deprived conditions through investigating the relationship between the glucose consumption rate and intracellular NADH/NAD+ ratio in various mutant strains. The NADH/NAD+ ratio was strongly repressed under oxygen deprivation when glucose consumption was accelerated by the addition of pyruvate or sodium hydrogen carbonate. Overexpression of the ndh gene in the wild-type strain under oxygen deprivation decreased the NADH/NAD+ ratio from 0.32 to 0.13, whereas the glucose consumption rate increased by 27 %. Similarly, in phosphoenolpyruvate carboxylase gene (ppc)- or malate dehydrogenase gene (mdh)-deficient strains, overexpression of the ndh gene decreased the NADH/NAD+ ratio from 1.66 to 0.37 and 2.20 to 0.57, respectively, whereas the glucose consumption rate increased by 57 and 330 %, respectively. However, in a lactate dehydrogenase gene (L-ldhA)-deficient strain, although the NADH/NAD+ ratio decreased from 5.62 to 1.13, the glucose consumption rate was not markedly altered. In a tailored d-lactate-producing strain, which lacked ppc and L-ldhA genes, but expressed D-ldhA from Lactobacillus delbrueckii, overexpression of the ndh gene decreased the NADH/NAD+ ratio from 1.77 to 0.56, and increased the glucose consumption rate by 50 %. Overall, the glucose consumption rate was found to be inversely proportional to the NADH/NAD+ ratio in C. glutamicum cultured under oxygen deprivation. These findings could provide an option to increase the productivity of chemicals and fuels under oxygen deprivation.





Similar content being viewed by others
References
Bartek T, Blombach B, Lang S, Eikmanns BJ, Wiechert W, Oldiges M, Nöh K, Noack S (2011) Comparative 13C metabolic flux analysis of pyruvate dehydrogenase complex-deficient, l-valine-producing Corynebacterium glutamicum. Appl Environ Microbiol 77:6644–6652
Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ (2011) Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol 77:3300–3310
Danshina PV, Schmalhausen EV, Avetisyan AV, Muronetz VI (2001) Mildly oxidized glyceraldehyde-3-phosphate dehydroge-nase as a possible regulator of glycolysis. IUBMB Life 51:309–314
Dominguez H, Rollin C, Guyonvarch A, Guerquin-Kern JL, Cocaign-Bousquet M, Lindley ND (1998) Carbon-flux distribution in the central metabolic pathways of Corynebacterium glutamicum during growth on fructose. Eur J Biochem 254:96–102
Duggleby RG, Dennis DT (1974) Nicotin-amide adenine dinucleotide-specific glyceral-dehyde 3-phosphate dehydrogenase from Pisum sativum. Assay and steady state kinetics. J Biol Chem 249:167–174
Garrigues C, Loubiere P, Lindley ND, Cocaign-Bousquet M (1997) Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J Bacteriol 179:5282–5287
Hasegawa S, Uematsu K, Natsuma Y, Suda M, Hiraga K, Jojima T, Inui M, Yukawa H (2012) Improvement of the redox balance increases l-valine production by Corynebacterium glutamicum under oxygen deprivation conditions. Appl Environ Microbiol 78:865–875
Hasegawa S, Suda M, Uematsu K, Natsuma Y, Hiraga K, Jojima T, Inui M, Yukawa H (2013) Engineering of Corynebacterium glutamicum for high-yield l-valine production under oxygen deprivation conditions. Appl Environ Microbiol 79:1250–1257
Heux S, Cachon R, Dequin S (2006) Cofactor engineering in Saccharomyces cerevisiae: Expression of a H2O-forming NADH oxidase and impact on redox metabolism. Metab Eng 8:303–314
Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H (2004) Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol 7:182–196
Inui M, Suda M, Okino S, Nonaka H, Puskas LG, Vertès AA, Yukawa H (2007) Transcriptional profiling of Corynebacterium glutamicum metabolism during organic acid production under oxygen deprivation conditions. Microbiology 153:2491–2504
Jaworowski A, Mayo G, Shaw DC, Campbell HD, Young IG (1981) Characterization of the respiratory NADH dehydrogenase of Escherichia coli and reconstitution of NADH oxidase in ndh mutant membrane vesicles. Biochemistry 20:3621–3628
Kinoshita S (1985) Glutamic acid bacteria. In: Demain AL, Solomon NA (eds) Biology of industrial microorganisms. Benjamin Cummings, London, pp 115–146
Koebmann BJ, Solem C, Pedersen MB, Nilsson D, Jensen PR (2002) Expression of genes encoding F1-ATPase results in uncoupling of glycolysis from biomass production in Lactococcus lactis. Appl Environ Microbiol 68:4274–4282
Leif H, Sled VD, Ohnishi T, Weiss H, Friedrich T (1995) Isolation and characterization of the proton-translocating NADH: ubiquinone oxidoreductase from Escherichia coli. Eur J Biochem 230:538–548
Liang L, Liu R, Wang G, Gou D, Ma J, Chen K, Jiang M, Wei P, Ouyang P (2012) Regulation of NAD(H) pool and NADH/NAD(+) ratio by overexpression of nicotinic acid phosphoribosyltransferase for succinic acid production in Escherichia coli NZN111. Enzyme Microb Technol 51:286–293
Litsanov B, Brocker M, Bott M (2012) Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl Environ Microbiol 78:3325–3337
Lopez de Felipe F, Kleerebezem M, de Vos WM, Hugenholtz J (1998) Cofactor engineering: a novel approach to metabolic engineering in Lactococcus lactis by controlled expression of NADH oxidase. J Bacteriol 180:3804–3808
Matsushita K, Ohnishi T, Kaback HR (1987) NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry 26:7732–7737
Matsushita K, Otofuji A, Iwahashi M, Toyama H, Adachi O (2001) NADH dehydrogenase of Corynebacterium glutamicum. Purification of an NADH dehydrogenase II homolog able to oxidize NADPH. FEMS Microbiol Lett 204:271–276
Molenaar D, van der Rest ME, Drysch A, Yucel R (2000) Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Corynebacterium glutamicum. J Bacteriol 182:6884–6891
Nakayama K, Kitada S, Kinoshita S (1961) Studies on lysine fermentation I. The control mechanism on lysine accumulation by homoserine and threonine. J Gen Appl Microbiol 7:145–154
Nantapong N, Kugimiya Y, Toyama H, Adachi O, Matsushita K (2004) Effect of NADH dehydrogenase-disruption and over-expression on respiration-related metabolism in Corynebacterium glutamicum KY9714. Appl Microbiol Biotechnol 66:187–193
Nantapong N, Otofuji A, Migita CT, Adachi O, Toyama H, Matsushita K (2005) Electron transfer ability from NADH to menaquinone and from NADPH to oxygen of type II NADH dehydrogenase of Corynebacterium glutamicum. Biosci Biotechnol Biochem 69:149–159
Okino S, Inui M, Yukawa H (2005) Production of organic acids by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 68:475–480
Okino S, Noburyu R, Suda M, Jojima T, Inui M, Yukawa H (2008) An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol 81:459–464
Palmfeldt J, Paese M, Hahn-Hagerdal B, Van Niel EW (2004) The pool of ADP and ATP regulates anaerobic product formation in resting cells of Lactococcus lactis. Appl Environ Microbiol 70:5477–5484
Radoš D, Turner DL, Fonseca LL, Carvalho AL, Blombach B, Eikmanns BJ, Neves AR, Santos H (2014) Carbon flux analysis by 13C nuclear magnetic resonance to determine the effect of CO2 on anaerobic succinate production by Corynebacterium glutamicum. Appl Environ Microbiol 80:3015–3024
Sambrook J, Fritsh E, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Tsuchida Y, Kimura S, Suzuki N, Inui M, Yukawa H (2009) Characterization of a 24-kb plasmid pCGR2 newly isolated from Corynebacterium glutamicum. Appl Microbiol Biotechnol 87:1855–1866
Tsuge Y, Yamamoto S, Suda M, Inui M, Yukawa H (2013) Reactions upstream of glycerate-1,3-bisphosphate drive Corynebacterium glutamicum d-lactate productivity under oxygen deprivation. Appl Microbiol Biotechnol 97:6693–6703
Yamamoto S, Gunji W, Suzuki H, Toda H, Suda M, Jojima T, Inui M, Yukawa H (2012) Overexpression of genes encoding glycolytic enzymes in Corynebacterium glutamicum enhances glucose metabolism and alanine production under oxygen deprivation conditions. Appl Environ Microbiol 78:4447–4457
Yamamoto S, Suda M, Niimi S, Inui M, Yukawa H (2013) Strain optimization for efficient isobutanol production using Corynebacterium glutamicum under oxygen deprivation. Biotechnol Bioeng 110:2938–2948
Yukawa H, Omumasaba CA, Nonaka H, Kos P, Okai N, Suzuki N, Suda M, Tsuge Y, Watanabe J, Ikeda Y, Vertès AA, Inui M (2007) Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 153:1042–1058
Acknowledgments
This work was supported by a grant from the New Energy and Industrial Technology Development Organization (NEDO). This study was also partially supported by a Grant-in-Aid for Young Scientists (B) to YT from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Conflict of interest
We declare that we have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tsuge, Y., Uematsu, K., Yamamoto, S. et al. Glucose consumption rate critically depends on redox state in Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 99, 5573–5582 (2015). https://doi.org/10.1007/s00253-015-6540-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6540-2