Abstract
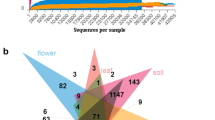
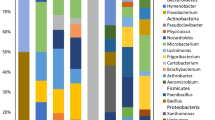
Plants harbors complex and variable microbial communities. Endophytic bacteria play an important function and potential role more effectively in developing sustainable systems of crop production. To examine how endophytic bacteria in sugar beet (Beta vulgaris L.) vary across both host growth period and location, PCR-based Illumina was applied to revealed the diversity and stability of endophytic bacteria in sugar beet on the north slope of Tianshan mountain, China. A total of 60.84 M effective sequences of 16S rRNA gene V3 region were obtained from sugar beet samples. These sequences revealed huge amount of operational taxonomic units (OTUs) in sugar beet, that is, 19–121 OTUs in a beet sample, at 3 % cutoff level and sequencing depth of 30,000 sequences. We identified 13 classes from the resulting 449,585 sequences. Alphaproteobacteria were the dominant class in all sugar beets, followed by Acidobacteria, Gemmatimonadetes and Actinobacteria. A marked difference in the diversity of endophytic bacteria in sugar beet for different growth periods was evident. The greatest number of OTUs was detected during rossette formation (109 OTUs) and tuber growth (146 OTUs). Endophytic bacteria diversity was reduced during seedling growth (66 OTUs) and sucrose accumulation (95 OTUs). Forty-three OTUs were common to all four periods. There were more tags of Alphaproteobacteria and Gammaproteobacteria in Shihezi than in Changji. The dynamics of endophytic bacteria communities were influenced by plant genotype and plant growth stage. To the best of our knowledge, this study is the first application of PCR-based Illumina pyrosequencing to characterize and compare multiple sugar beet samples.









Similar content being viewed by others
References
Adams PD, Kloepper JW (2002) Effect of host genotype on indigenous bacterial endophytes of cotton (Gossypium hirsutum L.). Plant Soil 240:181–189
Botella L, Santamaría O, Diez JJ (2010) Fungi associated with the decline of Pinus halepensis in Spain. Fungal Divers 40:1–11
Carroll GC (1995) Forest endophytes: pattern and process. Can J Bot 73:1316–1324
Chen HB (2012) Venn Diagram: generate high-resolution Venn and Euler plots. Rpackage version 113
Cocolin L, Rantsiou K, Iacumin L, Cantoni C, Comi G (2002) Direct identification in food samples of Listeria spp. and Listeria monocytogenes by molecular methods. Appl Environ Microbiol 68:6273–6282
Collado J, Platas G, Paleaz F (2001) Identification of an endophytic Nodulisporium sp. from Quercus ilexin central Spain as the anamorph of Biscogniauxia mediterranea by rDNA sequence analysis and effect of different ecological factors on distribution of the fungus. Mycologia 93:875–886
Cottrell MT, Kirchman DL (2000) Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl Environ Microbiol 66:5116–5122
Dunbar J, Takala S, Barns SM, Davis JA, Kuske CR (1999) Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl Environ Microbiol 65:1662–1669
Fisher PJ, Anson AE, Petrini O (1986) Fungal endophytes in Ulex europaeus and U. galli. Trans Br Mycol Soc 86:153–156
Fisher PJ, Petrini O (1990) A comparative study of fungal endophytes in xylem and bark of Alnus sp. in England and Switzerland. Mycol Res 94:313–319
Fisher PJ, Petrini O, Petrini LE, Sutton BC (1994) Fungal endophytes from the leaves and twigs of Quercus ilex L. from England, Majorca and Switzerland. New Phytol 127:133–137
Foissner W (1999) Protist diversity: estimates of the near-imponderable. Protist 150:363–368
Foissner W (2008) Protist diversity and distribution: some basic considerations. Biodivers Conserv 17:235–242
Gardner JM, Feldman AW, Zablotowicz M (1982) Identity and behaviour of xylem-residing bacteria in rough lemon roots of Florida citrus trees. Appl Environ Microbiol 43:1335–1342
Gloor GB, Hummelen R, Macklaim JM, Dickson RJ, Fernandes AD, MacPhee R, Reid G (2010) Microbiome profiling by Illumina sequencing of combinatorial sequence-tagged PCR products. PLoS One 5:e15406
Guo LD, Huang G, Wang Y (2008) Seasonal and tissue age influences on endophytic fungi of Pinus tabulaeformis (Pinaceae) in the Dongling Mountains, Beijing. J Integr Plant Biol 50:997–1003
Hallmann J, Quadt-Hallmann A, Mahafee WF, Kloepper JW (1997) Bacterial endophytes in agricultural crops. Can J Microbiol 43:895–914
Helander M, Wali P, Kuuluvainen T, Saikkonen K (2006) Birch leaf endophytes in managed and natural boreal forests. Can J For Res 36:3239–3245
Huse SM, Dethlefsen L, Huber JA, Mark WD, Relman DA, Sogin ML (2008) Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet 4:e1000255
Jacobs MJ, William MB, David AG (1985) Enumeration, location, characterization of endophytic bacteria within sugar beet roots. Can J Bot 63:1262–1265
Johnson JA, Whitney NJ (1994) Cytotoxicity and insecticidal activity of endophytic fungi from Black Spruce (Picea mariana) needles. Can J Microbiol 40:24–27
Kauhanen M., Vainio EJ, Hantula J, Eyjolfsdottir GG, Niemelä P (2006) Endophytic fungi in Siberian larch (Larix sibirica) needles. Forest Pathology 36:434–446
Kibe R, Sakamoto M, Yokota H, Ishikawa H, Aiba Y, Koga Y, Benno Y (2005) Movement and fixation of intestinal microbiota after administration of human feces to germfree mice. Appl Environ Microbiol 71:3171–3178
Kobayashi DY, Palumbo JD (2000) Bacterial endophytes and their effects on plants and uses in agriculture. In CW Bacon, JF White, eds, Microbial Endophytes. CRC Press, New York, pp 199–229
Kolde R (2012) pheatmap: Pretty Heatmaps. Rpackage version 061
Lazarevic V, Whiteson K, Huse S, Hernandez D, Farinelli L, Osteras M, Schrenzel J, François P (2009) Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Methods 79:266–271
Legault D, Dessureault M, Laflamme G (1989) Mycoflora of Pinus banksiana and Pinus resinosa needles. II. Epiphytic fungi. Can J Bot 67:2061–2065
Lodewyck C, Vangronsveld J, Porteous F, Moore ERB, Taghavi S, Mezgeay M, van der Lelie D (2002) Endophytic bacteria and their potential application. Crit Rev Plant Sci 86(6):583–606
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488(7409):86–90
Martín-García J, Espiga E, Pando V, Diez JJ (2011) Factors infuencing endophytic communities in poplar plantations. Silva Fennica 45:169–180
Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JH, Piceno YM, DeSantis TZ, Andersen GL, Bakker PA, Raaijmakers JM (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332(6033):1097–1100
Moore FP, Barac T, Borremans B, Oeyen L, Vangronsveld J, van der Lelie D, Campbell CD, Moore ER (2006) Endophytic bacterial diversity in poplar trees growing on a BTEX-contaminated site: the characterisation of isolates with potential to enhance phytoremediation. Syst Appl Microbiol 29:539–556
Pan MJ, Rademan S, Kuner K, Hastings JW (1997) Ultrastructural studies on the colonisation of banana tissue and Fusarium oxysporum f. sp. cubenserace 4 by the endophytic bacterium Burkholderia cepacia. J Phytopathol 145:479–486
Paul FK, Josephine YA (2004) Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol Ecol 47:161–177
Petrini O, Carroll GC (1981) Endophytic fungi in foliage of some Cupressaceae in Oregon. Can J Bot 59:629–636
Rademaker JLW, Louws FJ, de Bruijn FJ (1998) Characterisation of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting. Molecular microbial ecology manual. Kluwer Academic Publishers, Netherlands, pp 1–27
Riesenfeld CS, Schloss PD, Handelsman J (2004) Metagenomics: genomic analysis of microbial communities. Annu Rev Genet 38:525–552
Rodrigues KF (1994) The foliar fungal endophytes of the Amazonian palm Euterpe oleracea. Mycologia 86:376–385
Roll Hansen F, Roll Hansen H (1979) Ascocoryne species in living stems of Picea spp. Eur J For Pathol 5:275–280
Ryan RP, Germaine K, Franks A, Ryan DJ, Dowling DN (2008) Bacterial endophytes: recent developments and applications. FEMS Microbiol Lett 278:1–9
Samish Z, Etinger-Tulczynska R, Bick M (1963) The microflora within the tissue of soft fruit and vegetables. J Food Sci 28:259–266
Shi YW, Lou K, Li C (2009) Promotion of plant growth by phytohormone producing endophytic microbes of sugar beet. Biol Fertil Soils 45:645–653
Shi YW, Lou K, Li C (2010) Growth and photosynthetic efficiency promotion of sugar beet (Beta vulgaris L.) by endophytic bacteria. Photosynth Res 105:5–13
Shi YW, Lou K, Li C (2011) Growth promotion effects of the endophyte Acinetobacter johnsonii strain 3-1 on sugar beet. Symbiosis 54(3):159–166
Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, Roskot N, Heuer H, Berg G (2001) Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol 67:4742–4751
Smith DR, Quinlan AR, Peckham HE, Makowsky K, Tao W, Woolf B, Shen L, Donahue WF, Tusneem N, Stromberg MP, Stewart DA, Zhang L, Ranade SS, Warner JB, Lee CC, Coleman BE, Zhang Z, McLaughlin SF, Malek JA, Sorenson JM, Blanchard AP, Chapman J, Hillman D, Chen F, Rokhsar DS, McKernan KJ, Jeffries TW, Marth GT, Richardson PM (2008) Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res 18:1638–1642
Spear GT, Sikaroodi M, Zariffard MR, Landay AL, French AL, Gillevet PM (2008) Comparison of the diversity of the vaginal microbiota in HIV-infected and HIV-uninfected women with or without bacterial vaginosis. J Infect Dis 198:1131–1140
Spiers AJ, Buckling A, Raineythen PB (2000) The causes QJ; of Pseudomonas diversity. Microbiology 146:2345–2350
Sprent JI, de Faria SM (1988) Mechanisms of infection of plants by nitrogen fixing organisms. Plant Soil 110:157–165
Sun L, Qiu F, Zhang X, Dai X, Dong X, Song W (2008) Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microb Ecol 55:415–424
Tanprasert P, Reed BM (1997) Detection and identification of bacterial contaminants from strawberry runner explants. In Vitro Cell Dev Biol 33:221–226
Tauxe RV, Hargrett-Bean N, Patton CM, Wachsmuth IK (1988) Campylobacter isolates in the United States, 1982–1986. Morbid Mortal Wkly Rep 37:1–13
Tringe SG, Rubin EM (2005) Metagenomics: DNA sequencing of environmental samples. Nat Rev Genet 6:805–814
von Mering C, Hugenholtz P, Raes J, Tringe SG, Doerks T, Jensen LJ, Ward N, Bork P (2007) Quantitative phylogenetic assessment of microbial communities in diverse environments. Science 315:1126–1130
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Project No. 31060018).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shi, Y., Yang, H., Zhang, T. et al. Illumina-based analysis of endophytic bacterial diversity and space-time dynamics in sugar beet on the north slope of Tianshan mountain. Appl Microbiol Biotechnol 98, 6375–6385 (2014). https://doi.org/10.1007/s00253-014-5720-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5720-9