Abstract
The major murein and pseudomurein cell wall-binding domains, i.e., the Lysin Motif (LysM) (Pfam PF01476) and pseudomurein cell wall-binding (PMB) (Pfam PF09373) motif, respectively, were genetically fused. The fusion protein is capable of binding to both murein- and pseudomurein-containing cell walls. In addition, it also binds to chitin, the major polymer of fungal cell walls. Binding is influenced by pH and occurs at a pH close to the pI of the binding protein. Functional studies on truncated versions of the fusion protein revealed that murein and chitin binding is provided by the LysM domain, while binding to pseudomurein is achieved through the PMB domain.
Similar content being viewed by others
Introduction
Cells of most Gram-positive bacteria and methanogenic archaea of the orders Methanobacteriales and Methanopyrales are surrounded by a protective cell wall made of murein or pseudomurein, respectively (Kandler and König 1978, 1998; Steenbakkers et al. 2006; Visweswaran et al. 2010). Although murein and pseudomurein perform a similar function in these organisms, some fundamental differences exist in their composition and architecture. Murein is made of polymers of β(1-4) linked N-acetylmuramic acid and N-acetyl-d-glucosamine (NAG), whereas pseudomurein is composed of chains of β(1,3)-linked NAG and N-acetyltalosaminuronic acid residues. Murein and pseudomurein are substrates for a variety of cell wall hydrolases involved in various physiological processes like cell separation and autolysis. To facilitate in these biological processes, most cell wall hydrolases are equipped with one or more cell wall-binding domains that non-covalently attach the enzymes to their polymeric substrates.
In this study, a fusion protein was generated by the union of two cell wall-binding domains, i.e., the Lysin Motif (LysM) domain, which binds to bacterial cell walls, and the pseudomurein cell wall-binding (PMB) domain, which binds to pseudomurein-containing archaeal cell walls. The LysM domain is a ubiquitous, murein-binding domain present in proteins of both prokaryotes and eukaryotes (Bateman and Bycroft 2000; Buist et al. 2008). It has a wide range of possible applications both in industry and in medicine (Audouy et al. 2006; Bosma et al. 2006; Audouy et al. 2007). For instance, LysM domains are currently employed in the development of pneumococcal and influenza vaccines (Audouy et al. 2006, 2007; Bosma et al. 2006; Saluja et al. 2010). They are also used for the display of various heterologous proteins on the bacterial cell surface (Steen et al. 2003; Bosma et al. 2006; Okano et al. 2008; Tarahomjoo et al. 2008; Hu et al. 2010). In bacteria, LysM domains are present in murein hydrolases helping in the non-covalent attachment of the enzymes to the murein layer, thus facilitating substrate hydrolysis. Functional studies suggest that LysM domains recognize the NAG moiety of murein (Ohnuma et al. 2008; Petutschnig et al. 2010). In plants, LysM domains play a vital role in the symbiotic relationship between plants and some of their bacterial hosts (Mulder et al. 2006; Radutoiu et al. 2007; Wan et al. 2008). Plant LysM domains may also bind to chitin, the cell wall polymer of fungi, as part of the plants’ defense mechanism against pathogenic fungi (Ohnuma et al. 2008; Petutschnig et al. 2010).
On the other hand, the PMB domain is narrowly distributed and present only in a few methanogenic archaeal proteins and in two archaea-specific viral hydrolases (PeiP and PeiW) (Kiener et al. 1987; Stax et al. 1992; Pfister et al. 1998; Luo et al. 2001, 2002; Steenbakkers et al. 2006; Visweswaran et al. 2010, 2011a, b). Until now, there are no experimental reports demonstrating the occurrence of LysM domains in archaea (Buist et al. 2008) or PMB domains in bacteria. The function of the PMB domain in archaeal hydrolases is analogous to that of the LysM domains in bacterial hydrolases, i.e., binding of the enzyme to its polymeric substrate, in this case the pseudomurein layer in methanogenic archaeal cell walls (Visweswaran et al. 2010, 2011a, b). Deletion of the PMB domain in one of the pseudomurein endoisopeptidases, PeiW, led to loss of binding of the enzyme to the host cell wall (Steenbakkers et al. 2006). In analogy to the use of the LysM domain, the PMB domain might be employed for the surface display of heterologous proteins on pseudomurein-containing cell walls.
To obtain the combined properties of both the LysM and PMB domains and also to increase the number of binding partners, we developed a fusion protein by genetic fusion of the three C-terminal LysM motifs of a major autolysin, AcmA from Lactococcus lactis, and three C-terminal PMB motifs of the surface S-layer protein MTH719 from Methanothermobacter thermautotrophicus (see Fig. 1). The genetically engineered fusion protein was investigated with respect to binding to bacterial and methanogenic archaeal cell walls and the effect of pH thereon. Interestingly, the fusion protein was shown to bind to chitin flakes and to fungal cell walls through its LysM domain.
Molecular architecture and binding activity of the LysMAcmA, PMBMTH719, and the M–P–GFP–H10 fusion proteins. The three LysM motifs of AcmA and three PMB motifs of MTH719 are abbreviated as L1, L2, and L3 and P1, P2, and P3, respectively. GFP and H10 indicate the green fluorescent protein and the His10 tag located at the C-terminus, respectively. The binding activity of the different domain constructs to the cell wall materials is indicated with + (bound) or – (not bound)
Materials and methods
Strains and plasmids
The LysMAcmA, PMBMTH719, and the M–P fusion proteins used in this study were constructed in pBADcLIC-GFP plasmid, bearing an arabinose-inducible promoter, using the ligation-independent cloning method described previously (Geertsma and Poolman 2007). Oligos used for the amplification of specific DNA fragments are shown in Table 1. Primers LR and PF contain homologous nucleotide sequences to the PMBMTH719 domain and the LysMAcmA domain, respectively, to facilitate overlap PCR. Plasmids annealed with DNA fragments of individual constructs of LysMAcmA, PMBMTH719 domains, and the M–P fusion protein were used for transformation of Escherichia coli Rosetta gami 2 (Novagen, Darmstadt, Germany) using the heat shock method (van Die et al. 1983). Transformed cells were plated on selective TY 1.5% (w/v) agar plates and transformants were checked by colony PCR and plasmid DNA sequencing (ServiceXS, Leiden, the Netherlands) for the correct sequence and right orientation of the desired gene products. For overexpression of specific constructs, E. coli cells were grown with shaking at 37°C in TY broth (Difco, Sparks, MD, USA), containing ampicillin (50 μg/ml) or chloroamphenicol (50 μg/ml), as required. The murein-lacking planctomycetes cells (Rhodopirellula baltica) were grown on M607 agar plates; Sulfolobus acidocaldarius cells were grown on Brocks medium, supplemented with 0.2% (w/v) tryptone.
Protein expression, isolation, and purification
Fresh cultures of E. coli Rosetta gami 2 bearing the constructs of C-terminally GFP- and H10-tagged LysMAcmA domain, PMBMTH719 domain, and the M–P fusion protein were started from the mother cultures. Cells were grown at 37°C in selective TY medium until an OD at 600 nm of 0.6–0.8 was reached and were induced with 0.2% (w/v) arabinose (Merck KGaA, Darmstadt, Germany) followed by incubation for another 2 h at 37°C on a rotating shaker at 250 rpm (Innova 4000, New Brunswick Scientific, Edison, NJ, USA). The overexpressed proteins were isolated and purified separately according to the manufacturer’s protocol (Ni-NTA superflow, Qiagen GmbH, Hilden, Germany). GFP (control) was purified by hydrophobic interaction chromatography (HIC) according to the Bio-Rad experimental protocol.
In-gel fluorescence and western hybridization
The LysMAcmA, PMBMTH719, and M–P fusion protein bearing the C-terminal GFP and H10 tag were subjected to SDS 12.5%-PAGE and in-gel GFP fluorescence was visualized using a Gel Documentation System (Bio-Rad Laboratories Inc, Hercules, CA, USA). The same gel was blot-transferred onto polyvinylidene fluoride transfer membrane (GE Healthcare UK Limited, Little Chalfont, Buckinghamshire, UK). Anti-His-tag polyclonal rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and anti-rabbit IgG peroxidase (GE Healthcare UK Limited) were used as primary and secondary antibodies, respectively. Washing and incubation steps were performed using a SNAP-id system (Millipore). Enhanced chemiluminescence was used to detect the signals, according to the manufacturer’s protocol (GE Healthcare UK Limited).
Binding studies
Preparation of L. lactis Gram-positive Enhancer Matrix (GEM) particles containing purified murein was previously explained (Audouy et al. 2006, 2007; Bosma et al. 2006). Ni-NTA-purified LysMAcmA, PMBMTH719, and M–P fusion protein and HIC-purified control GFP protein were incubated separately with the L. lactis GEM particles, pseudomurein of Methanobacterium sp. cells (Sigma, Zwijndrecht, the Netherlands) chitin from shrimp shells (Sigma) and fungal cells (Psilocybe cubensis taxid: 181762) in 50 mM NaHCO3, pH 9.2 and 10.0 buffers. Additionally, the M–P–GFP–H10 fusion protein, purified GFP (control), and the substrates were incubated in 50 mM NaH2PO4, pH 6.0 and 8.0 buffers. As negative controls, cells of R. baltica, a murein-lacking bacterium, and S. acidocaldarius, an archaeon deficient in pseudomurein, were used in all the binding experiments. The incubation was performed at room temperature for 30 min on a rotatory shaker. The mixture was spun down at 14,000 rpm on a desktop centrifuge. The pellet was washed three times with the same buffer supplemented with 150 mM NaCl followed by centrifugation after every wash. After the incubation and washing steps, the material was resuspended in the same buffers. The material was placed on a microscope slide and viewed under a phase-contrast microscope (Zeiss Axiophot, Thornwood, USA) fitted with a digital camera and a green filter to view fluorescence. All photographs were taken at 1,250-fold magnification.
Results
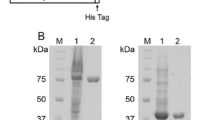
Nucleotide sequences encoding the LysM domain from the autolysin AcmA of L. lactis subsp. cremoris MG1363 (plasmid-free derivative of strain NCDO712 of the National Collection of Dairy Organisms) (Buist et al. 1995) and the PMB domain from the S-layer protein MTH719 of M. thermautotrophicus str. ∆H (strain DSMZ1053 of the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH) (Visweswaran et al. 2011a) were amplified separately by PCR using the primer pair LF/LR and PF/PR, respectively (see Table 1). The individual LysM and PMB amplification products were used as templates for the generation of the fusion gene by overlap PCR with the primer pair LF/PR. The three amplified DNA fragments were each cloned into pBADcLIC-GFP (Geertsma and Poolman 2007) using E. coli Rosetta gami 2. In this way, each protein domain was fused to a GFP and H10 tag. The molecular architecture of the individual LysM, PMB, and LysM–PMB fusion protein (from now onwards denoted as M–P) domains is shown in Fig. 1. The expression of the AcmA LysM (LysMAcmA), MTH719 PMB (PMBMTH719), and M–P fusion domains and their proper fusion to the C-terminal GFP–H10 tag, was confirmed by in-gel fluorescence detection and western hybridization using anti-His-tag antibodies (Fig. 2). The extra band in lane 5 (Fig. 2a) is most likely caused by protein degradation, liberating GFP, as the protein in this band also fluoresces (Fig. 2b). A non-fluorescent breakdown product can be seen in lane 3 (Fig. 2a, b).
Expression of M–P, LysMAcmA, and PMBMTH719 GFP–H10 fusion protein constructs. a Western blot using anti-His antibodies and b in-gel fluorescence. Lane 1, molecular mass marker; lanes 2, 4, and 6 and 3, 5, and 7, uninduced and 0.2% L-arabinose-induced cell free extracts of M–P–GFP–H10 fusion protein; LysMAcmA–GFP–H10 and PMBMTH719–GFP–H10, respectively. Arrows (↓) indicate specific protein band
Homologies of LysM and PMB motifs
LysM motifs contain 40–65 amino acids, whereas PMB motifs are 30–35 residues in length (Fig. 3) (Buist et al. 2008; Visweswaran et al. 2010, 2011a, b). No significant homology exists between the LysM and the PMB motifs. The three C-terminal LysM motifs of AcmA share homology with one another (Buist et al. 1995). A multiple sequence alignment using the ClustalW2 program (EMBL-EBI, Chenna et al. 2003) reveals 16 amino acid residues that are identical in all three motifs (Fig. 3a). Six of these amino acid residues tend to be highly conserved in most of the known LysM sequences including those of the LysMs of which 3D structures are known (MltD, PDB: 1E0G, and YkuD, PDB: 1Y7M) (Fig. 3a).
Multiple sequence alignments of the AcmA LysM and MTH719 PMB motifs using ClustalW2 (EMBL-EBI, Chenna et al. 2003). a The three C-terminal LysM motifs of the AcmA protein. b The three C-terminal PMB motifs of the MTH719 protein. Bold residues indicate highly conserved amino acids in consensus sequences (Bateman and Bycroft 2000) and asterisks (*) indicate identical residues in the three motifs. Amino acid residues common between MTH719 PMB repeats 1 and 3, 2 and 3, and 1 and 2 are indicated in blue, red, and green, respectively
In contrast, the three PMB motifs in PMBMTH719 are poorly conserved. Only two amino acid residues (R26, P28) are commonly found in most other PMB motif sequences (Fig. 3b and Pfam, http://pfam.sanger.ac.uk/family/pmbr#tabview=tab4). However, motifs 1 and 3 and motifs 2 and 3 possess 11 and nine identical amino acid residues between them, respectively, while motifs 1 and 2 have only four residues in common (Fig. 3b). The significance of the three PMB motifs of the MTH719 protein is that they contribute an overall positive charge to MTH719. The pI of PMBMTH719 is 10.6 while the pI of the MTH719 protein without the PMB domain is only 4.5 (ExPASy). Because of this high pI value of the PMB domain, the MTH719 protein as a whole possesses a relatively high pI (8.7) (ExPASy). In contrast, the pI of the AcmA protein is not significantly affected by the presence of the LysM domain.
The LysM and PMB domains specifically bind to their respective substrates
To demonstrate the functionality of the individual domains in the LysMAcmA–GFP–H10 and PMBMTH719–GFP–H10 proteins, both were purified using Ni-NTA column chromatography. LysMAcmA–GFP–H10 was incubated with L. lactis GEM particles (Audouy et al. 2006, 2007; Bosma et al. 2006), shrimp shell chitin flakes, pseudomurein of Methanobacterium sp. cells, and cells of P. cubensis at pH 10.0. LysMAcmA–GFP–H10, specifically bound to the murein-containing L. lactis GEM particles (Figs. 1 and 4a). Interestingly, it also bound to shrimp shell chitin flakes and to the chitin-containing fungal (P. cubensis) cell walls (Figs. 1 and 4b, c). No binding was observed to the pseudomurein of Methanobacterium sp. cells (Figs. 1 and 4a). HIC-purified GFP was used as a negative control in the binding experiments and did not give any fluorescence signal with all tested substrates (data not shown). Additionally, cells of R. baltica and S. acidocaldarius were used as murein and pseudomurein-deficient controls, respectively. No protein binding was observed (data not shown). This indicates that the LysMAcmA domain is responsible for binding to the three substrates (L. lactis GEM particles, shrimp shell chitin flakes, and cells of P. cubensis) and shows, for the first time, that a LysM domain from a Gram-positive bacterium can also bind chitin, as can their plant counterparts (Ohnuma et al. 2008; Petutschnig et al. 2010).
Binding specificity of the LysMAcmA–GFP–H10 and PMBMTH719–GFP–H10 proteins. The figures (a–c) show fluorescence microscopy views of the LysMAcmA–GFP–H10 binding selectively and specifically to L. lactis GEM particles (cocci), chitin flakes, and P. cubensis cells, respectively, at pH 10.0. The figure (d) shows binding specificity of PMBMTH719–GFP–H10 protein to pseudomurein-containing Methanobacterium sp. cells (long rods), at pH 10.0
When purified PMBMTH719–GFP–H10 was incubated with the same substrates at pH 10.0, binding was only observed with pseudomurein-containing Methanobacterium sp. cells (Figs. 1 and 4d); no GFP fluorescence was seen with the other substrates (Fig. 4d and data not shown) indicating the specificity of the PMBMTH719 domain towards pseudomurein-containing Methanobacterium sp. cells.
The M–P fusion protein is capable of binding to murein, pseudomurein, and chitin
Ni-NTA-purified M–P–GFP–H10 fusion protein was incubated at pH 10.0 with the same substrates tested earlier for the individual LysMAcmA and PMBMTH719 domains. The fusion protein bound to both whole cells of L. lactis, to GEM particles, and to pseudomurein of Methanobacterium sp. cells at room temperature (Figs. 1 and 5a). The M–P–GFP–H10 fusion protein also bound to shrimp shell chitin flakes and to P. cubensis fungal cell walls (Figs. 1 and 5b, c). In contrast, the M–P–GFP–H10 fusion protein did not bind to the negative controls (cells of R. baltica and S. acidocaldarius) (Fig. 5a). As HIC-purified GFP protein (control) did not give a fluorescence signal with the substrates (data not shown), the binding of the M–P–GFP–H10 fusion protein was specific.
Multiple substrate-binding ability of M–P–GFP–H10 fusion protein. The figure shows fluorescence microscopy views of M–P–GFP–H10 fusion protein binding at pH 10.0 to a L. lactis GEM particles (cocci) and pseudomurein-containing Methanobacterium sp. cells (long rods). Cells of R. baltica (round phase—dark, arrows) and S. acidocaldarius (round phase—gray, arrow heads) show no binding to b chitin flakes and c P. cubensis cells
Binding is pH dependent
Different pH conditions were tested to study the effect of pH on binding of the M–P–GFP–H10 fusion protein. To rule out the influence of pH on the GFP signal, HIC-purified GFP was tested at different pH prior to the binding experiments. Under all pH conditions tested, the GFP signal remained the same. The Ni-NTA-purified M–P–GFP–H10 fusion protein (pI 10.3) was mixed with L. lactis GEM particles, pseudomurein of Methanobacterium sp. cells, chitin flakes, and cells of the fungus P. cubensis in buffers of pH 6.0, 8.0, 9.6, or 10.0. No GFP signal was seen at pH 6.0 and 8.0, indicating that no binding occurred (data not shown). A very low GFP signal was detected at pH 9.6 (data not shown). At pH 10.0, a strong GFP signal was observed in all tested samples (Fig. 5), indicating that the M–P–GFP–H10 protein had bound to the four substrates.
We conclude that M–P fusion protein is capable of binding to multiple cell wall substrates due to the combined properties of the two major murein and pseudomurein cell wall-binding domains, i.e., LysMAcmA and PMBMTH719. We also showed that the LysMAcmA domain from bacterial origin can bind chitin.
Discussion
AcmA, the widely characterized autolysin of L. lactis, possesses an N-terminal catalytic glucosaminidase domain followed by a C-terminal LysM domain consisting of three motifs for binding to murein. Using LysMAcmA–GFP fusion studies, we have determined that the LysMAcmA domain binds to substrates other than murein. LysMAcmA did not only bind to murein-containing L. lactis GEM particles but also to chitin and to fungal cell walls. To date, only eukaryotic LysM domains were shown to bind to chitin (Ohnuma et al. 2008; Petutschnig et al. 2010). Here, we show for the first time the prokaryotic LysM domain’s ability to bind eukaryotic cell walls, i.e., chitin and to the cell wall of the fungus P. cubensis. The prokaryotic and eukaryotic LysM domains vary quite considerably in their secondary structures and mode of bonding. Unlike eukaryotic LysM domains, the LysM domains of prokaryotes do not possess disulfide bonds, rather the latter domains are supported by extensive secondary structure and hydrogen bonding (Buist et al. 2008). Furthermore, the amino acid sequences of LysM domains of chitinases and bacterial cell wall hydrolases are strikingly different (Steen et al. 2003).
NAG is the only common moiety between murein and chitin, and moreover, the mode of bonding is also the same, i.e., β(1,4). Previously, it was shown that LysM domains from plant origin bind to NAG derivatives (Ohnuma et al. 2008; Petutschnig et al. 2010). We presume that, like its eukaryotic counterparts, LysMAcmA might also recognize NAG in both murein and chitin. For a better understanding of LysM functionality and to investigate the nature of the substrate and the mode of binding, a ligand-bound structure of the LysM domain would be essential and an interesting target for future research.
Unlike the LysMAcmA domain, the PMBMTH719 domain specifically binds to pseudomurein-containing Methanobacterium sp. cells (Fig. 4d). This result is supported by studies on the PMB domain of PeiW, a pseudomurein endoisopeptidase containing four N-terminal PMB motifs, which showed that the protein specifically bound to pseudomurein-containing archaeal cells (Steenbakkers et al. 2006). Although the PMB domains of MTH719 and PeiW are functionally similar in specifically binding to pseudomurein, both domains have less than 20% amino acid sequence homology (data not shown). PMBMTH719 does not bind to chitin nor to intact murein, but it was recently shown that the PMB domains of both PeiW and MTH719 bind to bacterial spheroplasts (Visweswaran et al. 2011a). As these were made by lysozyme treatment of whole lactococcal cells, it was suggested that NAG is the binding ligand. The only common moiety in all three cell wall polymers (murein, pseudomurein and chitin) is NAG. Whether PMBMTH719 recognizes NAG or β(1,3)-linked NAG might be resolved by a 3D structure of the domain with its ligand.
The M–P–GFP–H10 fusion protein obtained by the union of the LysMAcmA domain and the PMBMTH719 domain was shown to be functionally active even after the genetic fusion of these two domains from rather distinct origins (bacteria and archaea) and despite the presence of the GFP–H10 tag on the C-terminus of the triple fusion. This indicates that the individual domains (LysMAcmA and PMBMTH719) in the M–P fusion protein are correctly folded. Due to their combined properties, M–P–GFP–H10 could bind to all the substrates tested (murein, pseudomurein, and chitin). Based on these results and together with previously obtained results from the individual domains, we conclude that binding of M–P–GFP–H10 to GEM particles, chitin flakes, and to P. cubensis fungal cell walls was mediated by the LysMAcmA domain, and binding to pseudomurein-containing Methanobacterium sp. cells is effectuated by the PMBMTH719 domain. The binding of M–P–GFP–H10 is greatly influenced by the pH of the binding environment and depends on the individual pI values of the LysMAcmA and PMBMTH719 domains. Previous work from our group on the individual LysMAcmA (pI 10.0) and PMBMTH719 (pI 10.6) domains has shown that both bind to their substrates at a pH close to their respective pI values (Buist et al. 1995; Steen et al. 2003, 2005; Visweswaran et al. 2011a). The M–P–GFP–H10 fusion protein (pI 10.3) bound to substrates at a pH close to its pI, i.e., pH 10.0, indicating pH dependency. The LysM domain of AcmD, an N-acetylglucosaminidase from L. lactis and a homologue of AcmA, binds to L. lactis cells at pH of 4.0, which is close to the pI value of LysMAcmD (pI 4.3) (Visweswaran et al., unpublished results). An M–P–GFP–H10 fusion protein (pI 9.6) made of the LysMAcmD (pI 4.3) and PMBMTH719 (pI 10.6) bound to L. lactis GEM particles at a pH of 4.0 and to pseudomurein of Methanobacterium sp. cells at pH 10.0, but not to both substrates at an intermediate pH (pH 7.0), due to the quite different pI values of the two domains (data not shown).
In vivo, the prokaryotic LysM domains present in cell wall hydrolases aid in non-covalent binding of the enzyme to their substrates allowing some of the physiological functions like cell separation and cellular autolysis to work efficiently (Steen et al. 2005; Buist et al. 2008). Similarly, the PMB domains of archaeal hydrolases (PeiW and PeiP) allow binding of the enzymes to pseudomurein, facilitating hydrolysis by the catalytic domains (Luo et al. 2002; Steenbakkers et al. 2006; Visweswaran et al. 2010). In recent years, the LysM domains have been widely applied for various biotechnological purposes such as in the preparation of oral influenza vaccine and for surface display of heterologous proteins on bacterial cell surfaces (Bosma et al. 2006; Okano et al. 2008; Shao et al. 2009; Hu et al. 2010; Saluja et al. 2010; Xu et al. 2011). Similarly, the PMB domain could be employed for the display of target proteins on pseudomurein-containing archaeal cell surfaces; it could also be used as a marker protein to identify pseudomurein-containing methanogens from their counterparts. The fusion of the PMB domain to the LysM domain adds additional functionality to the latter, widening its application potential.
References
Audouy SA, van Roosmalen ML, Neef J, Kanninga R, Post E, van Deemter M, Metselaar H, van Selm S, Robillard GT, Leenhouts KJ, Hermans PW (2006) Lactococcus lactis GEM particles displaying pneumococcal antigens induce local and systemic immune responses following intranasal immunization. Vaccine 24:5434–5441
Audouy SA, van Selm S, van Roosmalen ML, Post E, Kanninga R, Neef J, Estevão S, Nieuwenhuis EE, Adrian PV, Leenhouts K, Hermans PW (2007) Development of lactococcal GEM-based pneumococcal vaccines. Vaccine 25:2497–2506
Bateman A, Bycroft M (2000) The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J Mol Biol 299:1113–1119
Bosma T, Kanninga R, Neef J, Audouy SAL, van Roosmalen ML, Steen A, Buist G, Kok J, Kuipers OP, Robillard G, Leenhouts K (2006) Novel surface display system for proteins on non-genetically modified Gram-positive bacteria. Appl Environ Microbiol 72:880–889
Buist G, Kok J, Leenhouts K, Dabrowska M, Venema G, Haandrikman A (1995) Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol 177:1554–1563
Buist G, Steen A, Kok J, Kuipers OP (2008) LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68:838–847
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500
Geertsma ER, Poolman B (2007) High-throughput cloning and expression in recalcitrant bacteria. Nat Meth 4:705–707
Hu S, Kong J, Kong W, Guo T, Ji M (2010) Characterization of a novel LysM domain from Lactobacillus fermentum bacteriophage endolysin and its use as an anchor to display heterologous proteins on the surfaces of lactic acid bacteria. Appl Environ Microbiol 76:2410–2418
Kandler O, König H (1978) Chemical composition of the peptidoglycan-free cell walls of methanogenic bacteria. Arch Microbiol 118(2):141–152
Kandler O, König H (1998) Cell wall polymers in Archaea (Archaebacteria). Cell Mol Life Sci 54(4):305–308
Kiener A, König H, Winter J, Leisinger T (1987) Purification and use of Methanobacterium wolfeii pseudomurein endopeptidase for lysis of Methanobacterium thermoautotrophicum. J Bacteriol 169:1010–1016
Luo Y, Pfister P, Leisinger T, Wasserfallen A (2001) The genome of archaeal prophage ΨM100 encodes the lytic enzyme responsible for autolysis of Methanothermobacter wolfeii. J Bacteriol 183:5788–5792
Luo Y, Pfister P, Leisinger T, Wasserfallen A (2002) Pseudomurein endoisopeptidases PeiW and PeiP, two moderately related members of a novel family of proteases produced in Methanothermobacter strains. FEMS Microbiol Lett 208:47–51
Mulder L, Lefebvre B, Cullimore J, Imberty A (2006) LysM domains of Medicago truncatula NFP protein involved in Nod factor perception. Glycosylation state, molecular modeling and docking of chitooligosaccharides and Nod factors. Glycobiol 16:801–809
Ohnuma T, Onaga S, Murata K, Taira T, Katoh E (2008) LysM domains from Pteris ryukyuensis chitinase-A. J Biol Chem 283:5178–5187
Okano K, Zhang Q, Kimura S, Narita J, Tanaka T, Fukuda H, Kondo A (2008) System using tandem repeats of the cA peptidoglycan-binding domain from Lactococcus lactis for display of both N- and C-terminal fusions on cell surfaces of lactic acid bacteria. Appl Environ Microbiol 74:1117–1123
Petutschnig EK, Jones AME, Serazetdinova L, Lipka U, Lipka V (2010) The Lysin Motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem 285:28902–28911
Pfister P, Wasserfallen A, Stettler R, Leisinger T (1998) Molecular analysis of Methanobacterium phage ΨM2. Mol Microbiol 30:233–244
Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J (2007) LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J 26:3923–3935
Saluja V, Amorij JP, Roosmalen ML, Leenhouts K, Huckriede A, Hinrichs WLJ, Frijlink HW (2010) Intranasal delivery of influenza subunit vaccine formulated with GEM particles as an adjuvant. AAPS J 12:109–116
Shao X, Jiang M, Yu Z, Cai H, Li L (2009) Surface display of heterologous proteins in Bacillus thuringiensis using a peptidoglycan hydrolase anchor. Microb Cell Fact 8:48
Stax D, Hermann R, Falchetto R, Leisinger T (1992) The lytic enzyme in bacteriophage ΨM1-induced lysates of Methanobacterium thermoautotrophicum Marburg. FEMS Microbiol Lett 100:433–438
Steen A, Buist G, Leenhouts KJ, Khattabi ME, Grijpstra F, Zomer AL, Venema G, Kuipers OP, Kok J (2003) Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J Biol Chem 278:23874–23881
Steen A, Buist G, Horsburgh GJ, Venema G, Kuipers OP, Foster SJ, Kok J (2005) AcmA of Lactococcus lactis is an N-acetylglucosaminidase with an optimal number of LysM domains for proper functioning. FEBS J 272:2854–2868
Steenbakkers PJM, Geerts WJ, Ayman-Oz NA, Keltjens JT (2006) Identification of pseudomurein cell wall binding domains. Mol Microbiol 62:1618–1630
Tarahomjoo S, Katakura Y, Shioya S (2008) Expression of C-terminal repeat region of peptidoglycan hydrolase of Lactococcus lactis IL1403 in methylotrophic yeast Pichia pastoris. J Biosci Bioeng 105:134–139
Van Die IM, Bergmans HEN, Hoekstra WPM (1983) Transformation in Escherichia coli: studies on the role of the heat shock in induction of competence. J Gen Microbiol 129:663–670
Visweswaran GRR, Dijkstra BW, Kok J (2010) Two major archaeal pseudomurein endoisopeptidases: PeiW and PeiP. Archaea. doi:10.1155/2010/480492
Visweswaran GRR, Dijkstra BW, Kok J (2011a) A minimum of three motifs is essential for optimal binding of pseudomurein cell wall-binding domain of Methanothermobacter thermautotrophicus. PLoS One 6(6):e21582. doi:10.1371/journal.pone.0021582
Visweswaran GRR, Dijkstra BW, Kok J (2011b) Murein and pseudomurein cell wall binding domains of bacteria and archaea—a comparative view. Appl Microbiol Biotechnol 92(5):921–928
Wan J, Zhang X, Neece D, Ramonell KM, Clough S, Kim S, Stacey MG, Stacey G (2008) A lysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20:471–481
Xu W, Huang M, Zhang Y, Yi X, Dong W, Gao X, Jia C (2011) Novel surface display system for heterogonous proteins on Lactobacillus plantarum. Lett Appl Microbiol 53(6):641–648. doi:10.1111/j.1472-765X.2011.03160.x
Acknowledgments
We thank Dr. J. Chong, University of York, for providing chromosomal DNA of M. thermautotrophicus. L. lactis GEM particles were a kind gift from K. Leenhouts and M. L. Roosmalen, Mucosis B.V, Groningen, the Netherlands. P. cubensis cells were a gift from the Department of Microbial Ecology, University of Groningen, the Netherlands. We are obliged to Dr. O. M. Lage, Department of Biology, University of Porto, Portugal and the Department of Molecular Microbiology, University of Groningen, the Netherlands for providing R. baltica and S. acidocaldarius cells, respectively.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Visweswaran, G.R.R., Dijkstra, B.W. & Kok, J. A genetically engineered protein domain binding to bacterial murein, archaeal pseudomurein, and fungal chitin cell wall material. Appl Microbiol Biotechnol 96, 729–737 (2012). https://doi.org/10.1007/s00253-012-3871-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-3871-0