Abstract
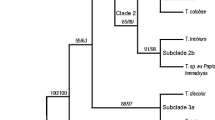
In a phylogenetic analysis of vertebrate transferrins (TFs), six major clades (subfamilies) were identified: (a) S, the mammalian serotransferrins; (b) ICA, the mammalian inhibitor of carbonic anhydrase (ICA) homologs; (c) L, the mammalian lactoferrins; (d) O, the ovotransferrins of birds and reptiles; (e) M, the melanotransferrins of bony fishes, amphibians, reptiles, birds, and mammals; and (f) M-like, a newly identified TF subfamily found in bony fishes, amphibians, reptiles, and birds. A phylogenetic tree based on the joint alignment of N-lobes and C-lobes supported the hypothesis that three separate events of internal duplication occurred in vertebrate TFs: (a) in the common ancestor of the M subfamily, (b) in the common ancestor of the M-like subfamily, and (c) in the common ancestor of other vertebrate TFs. The S, ICA, and L subfamilies were found only in placental mammals, and the phylogenetic analysis supported the hypothesis that these three subfamilies arose by gene duplication after the divergence of placental mammals from marsupials. The M-like subfamily was unusual in several respects, including the presence of a uniquely high proportion of clade-specific conserved residues, including distinctive but conserved residues in the sites homologous to those functioning in carbonate binding of human serotransferrin. The M-like family also showed an unusually high proportion of cationic residues in the positively charged region corresponding to human lactoferrampin, suggesting a distinctive role of this region in the M-like subfamily, perhaps in antimicrobial defense.





Similar content being viewed by others
References
Alemany R, Vilá MR, Franci C, Egea G, Real FX (1993) Glycosyl phosphatidylinositol membrane anchoring of melanotransferrin (p97): apical compartmentalization in intestinal epithelial cells. J Cell Sci 104:1155–1162
Ando K, Hasegawa K, Shindo K, Furusawa T, Fujino T, Kikugawa K, Nakano H, Takeuchi O, Akira S, Akiyama T, Gohda J, Inoue J, Hayakawa M (2010) Human lactoferrin activates NF-κB through the Toll-like receptor 4 pathway while it interferes with the lipopolysaccharide-stimulated TLR4 signaling. FEBS J 277:2051–2068
Baker EN, Baker HM (2009) A structural framework for understanding the multifunctional character of lactoferrin. Biochimie 91:3–10
Baker EN, Baker HM, Smith CA, Stebbins MR, Kahn M, Hellström KE, Hellström I (1992) Human melanotransferrin (p97) has only one functional iron-binding site. FEBS Lett 298:215–218
Baker EN, Baker HM, Kidd RD (2002) Lactoferrin and transferrin: functional variations on a common structural framework. Biochem Cell Biol 80:27–34
Bowman BH, Yang F, Adrian GS (1988) Transferrin: evolution and genetic regulation and expression. Adv Genet 25:1–38
Eckenroth BE, Mason AB, McDevitt ME, Lambert LA, Everse SJ (2010) The structure and evolution of the murine inhibitor of carbonic anhydrase: a member of the transferrin superfamily. Protein Sci 19:1616–1626
Escrivá H, Pierce A, Coddeville B, González F, Benaissa M, Léger D, Wieruszeski J-M, Spik G, Pamblanco M (1995) Rat mammary-gland transferrin: nucleotide sequence, phylogenetic analysis and glycan structure. Biochem J 367:47–55
Farnaud S, Evans RW (2003) Lactoferrin—a multifunctional protein with antimicrobial properties. Mol Immunol 40:395–400
García-Montoya IA, Cendón TS, Arévalo-Gallegos S, Rascón-Cruz Q (2012) Lactoferrin a multiple bioactive protein: an overview. Biochim Biophy Acta 1820:226–236
Giansanti F, Leboffe L, Pitari G, Ippoliti R, Antonini G (2012) Physiological roles of ovotransferrin. Biochim Biophys Acata 1820:218–225
Gkouvatsos K, Papanikolaou G, Pantopoulos K (2012) Regulation of iron transport and the role of transferrin. Biochim Biophys Acta 1820:188–202
Gomme PT, McCann KB (2005) Transferrin: structure, function and potential therapeutic actions. Drug Discov Today 10:267–273
Gray-Owen SD, Schyvers AB (1996) Bacterial transferrin and lactoferrin receptors. Trends Microbiol 4:185–191
Harris WR (2012) Anion binding properties of the transferrins. Implications for function. Biochim Biophys Acta 1820:348–361
Hedges SB (2009) Vetrebrates (Vertebrata). In: Hedges SB, Kumar S (eds) The timetree of life. Oxford University Press, Oxford, pp 309–314
Hughes AL (2014) Evolutionary diversification of aminopeptidase N in Lepidoptera by conserved derived amino acid residues. Mol Phyl Evol 76:127–133
Hunter HN, Demcoe AR, Jenssen H, Gutteberg TJ, Vogel HJ (2005) Human lactoferricin is partially folded in aqueous solution and is better stabilized in a membrane mimetic solvent. Antimicrob Agents Chemother 49:3387–3395
Jennssen H, Hancock RE (2009) Antimicrobial properties of lactoferrin. Biochimie 91:19–29
Kurokawa H, Mikami B, Hirose M (1995) Crystal structure of diferric hen ovotratransferrin at 2.4 Å resolution. J Mol Biol 254:196–207
Lambert LA (2012) Molecular evolution of the transferrin family and associated receptors. Biochim Biophy Acta 1820:244–255
Lambert LA, Perri H, Meehan TJ (2005) Evolution of duplications in the transferrin family of proteins. Comp Biochem Physiol B 140:11–25
Levay PF, Viljoen M (1995) Lactoferrin: a general review. Haematologica 80:252–267
Madsen O (2009) Mammals (Mammalia). In: Hedges SB, Kumar S (eds) The timetree of life. Oxford University Press, Oxford, pp 459–461
Mayle KM, Le AM, Kamei DT (2012) The intracellular trafficking pathway of transferrin. Biochim Biophys Acta 1820:264–281
Mizutani K, Toyoda M, Mikami B (2012) X-ray structures of transferrins and related proteins. Biochim Biophys Acta 1820:203–211
Murphy WJ, Eizirik E (2009) Placental mammals (Eutheria). In: Hedges SB, Kumar S (eds) The timetree of life. Oxford University Press, Oxford, pp 471–474
Nakamusu K, Kawamoto T, Shen M, Gotoh O, Teramoto M, Noshiro M, Kato Y (1999) Membrane-bound transferrin-like protein (MTf): structure, evolution and selective expression during chondrogenic differentiation of mouse embryonic cells. Biochim Biophys Acta 1447:258–264
Nibbering PH, Ravensbergen E, Welling MM, van Berkel LA, van Berkel PH, Pauwels EK, Nuijens JH (2001) Human lactoferrin and peptides derived from its N terminus are highly effective against interactions with antibiotic-resistant bacteria. Infect Immun 69:1469–1476
Pais FS-M, de Cássia Ruy P, Oliveira G, Coimbra RS (2014) Assessing the efficiency of multiple sequence alignment programs. Algorithms Mol Biol 9:4
Prasad AB, Allard MW, Comparative Sequencing Program NISC, Green ED (2008) Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol 25:1795–1808
Rahmanto YS, Bal S, Loh KH, Yu Y, Richardson DR (2012) Melanotransferrin: search for a function. Biochim Biophys Acta 1820:237–243
Sinha M, Kaushik S, Kaur P, Sharma S, Singh TP (2013) Antimicrobial lactoferrin peptides: the hidden players in the protective function of a multifunctional protein. Int J Peptides 2013:390230
Sun H, Li H, Sadler PJ (1999) Transferrin as a metal ion mediator. Chem Rev 99:2817–2842
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thorstensen K, Romslo I (1990) The role of transferrin in the mechanism of cellular iron uptake. Biochem J 271:1–10
Torda AE (2014) Not assessing the efficiency of multiple sequence alignment programs. Algorithm Mol Biol 9:18
Wang F, Lothrop AP, James NG, Griffiths TA, Lambert LA, Leverence R, Kaltashov IA, Andrews NC, MacGillivray RT, Mason AB (2007) A novel murine protein with no effect on iron homeostasis is homologous with transferrin and is the putative inhibitor of carbonic anhydrase. Biochem J 406:85–95
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hughes, A.L., Friedman, R. Evolutionary diversification of the vertebrate transferrin multi-gene family. Immunogenetics 66, 651–661 (2014). https://doi.org/10.1007/s00251-014-0798-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-014-0798-x