Abstract
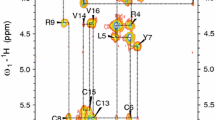
Protegrin pore formation is believed to occur in a stepwise fashion that begins with a nonspecific peptide interaction with the negatively charged bacterial cell walls via hydrophobic and positively charged amphipathic surfaces. There are five known nature protegrins (PG1-PG5), and early studies of PG-1 (PDB ID:1PG1) shown that it could form antiparallel dimer in membrane mimicking environment which could be a first step for further oligomeric membrane pore formation. Later, we solved PG-2 (PDB ID:2MUH) and PG-3 (PDB ID:2MZ6) structures in the same environment and for PG-3 observed a strong dαα NOE effects between residues R18 and F12, V14, and V16. These “inconsistent” with monomer structure NOEs appears due to formation of an additional antiparallel β-sheet between two monomers. It was also suggested that there is a possible association of protegrins dimers to form octameric or decameric β-barrels in an oligomer state. In order to investigate a more detailed oligomerization process of protegrins, in the present article we report the monomer (PDB ID: 2NC7) and octamer pore structures of the protegrin-5 (PG-5) in the presence of DPC micelles studied by solution NMR spectroscopy. In contrast to PG-1, PG-2, and PG-3 studies, for PG-5 we observed not only dimer NOEs but also several additional NOEs between side chains, which allows us to calculate an octamer pore structure of PG-5 that was in good agreement with previous AFM and PMF data.




Similar content being viewed by others
References
Aminova RM, Galiullina LF, Silkin NI, Ulmetov AR, Klochkov VV, Aganov AV (2013) Investigation of complex formation between hydroxyapatite and fragments of collagen by NMR spectroscopy and quantum-chemical modeling. J Mol Struct 1049:13–21
Ayupov RKh, Akberova NI (2015) Molecular dynamics of the pyridoxine derivative in the acetylcholinesterase active cavity. Res J Pharm Biol Chem Sci 6:1717–1722
Bessalle R, Kapitkovsky A, Gorea A, Shalit I, Fridkin M (1990) All-D-magainin—chirality, antimicrobial activity and proteolytic resistance. FEBS Lett 274:151–155
Bolintineanu DS, Kaznessis YN (2011) Computational studies of protegrin antimicrobial peptides: a review. Peptides 32:188–201
Borkar MR, Pissurlenkar RRS, Coutinho EC (2015) Mapping activity elements of protegrin antimicrobial peptides by HomoSAR. RSC Adv 5:78790–78798
Brogden KA (2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3:238–250
Capone R, Mustata M, Jang H, Arce FT, Nussinov R, Lal R (2010) Antimicrobial protegrin-1 forms ion channels: molecular dynamic simulation, atomic force microscopy, and electrical conductance studies. Biophys J 98:2644–2652
Chen J, Falla TJ, Liu HJ, Hurst MA, Fujii CA, Mosca DA, Embree JR, Loury DJ, Radel PA, Chang CC, Gu L, Fiddes JC (2000) Development of protegrins for the treatment and prevention of oral mucositis: structure-activity relationships of synthetic protegrin analogues. Biopolymers 55:88–98
Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D 66:12–21
Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) Nmrpipe—a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Efimov SV, Khodov IA, Ratkova EL, Kiselev MG, Berger S, Klochkov VV (2016) Detailed NOESY/T-ROESY analysis as an effective method for eliminating spin diffusion from 2D NOE spectra of small flexible molecules. J Mol Struct 1104:63–69
Epand RM, Vogel HJ (1999) Diversity of antimicrobial peptides and their mechanisms of action. BBA Biomembr 1462:11–28
Fahrner RL, Dieckmann T, Harwig SSL, Lehrer RI, Eisenberg D, Feigon J (1996) Solution structure of protegrin-1, a broad-spectrum antimicrobial peptide from porcine leukocytes. Chem Biol 3:543–550
Hancock REW (1997) Peptide antibiotics. Lancet 349:418–422
Hill CP, Yee J, Selsted ME, Eisenberg D (1991) Crystal-structure of defensin Hnp-3, an amphiphilic dimer—mechanisms of membrane permeabilization. Science 251:1481–1485
Hwang TL, Shaka AJ (1995) Water suppression that works—excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J Magn Reson Ser A 112:275–279
Jang H, Arce FT, Mustata M, Ramachandran S, Capone R, Nussinov R, Lal R (2011) Antimicrobial protegrin-1 forms amyloid-like fibrils with rapid kinetics suggesting a functional link. Biophys J 100:1775–1783
Khodov IA, Alper GA, Mamardashvili GM, Mamardashvili NZ (2015) Hybrid multi-porphyrin supramolecular assemblies: synthesis and structure elucidation by 2D DOSY NMR studies. J Mol Struct 1099:174–180
Khodov IA, Efimov SV, Klochkov VV, de Carvalho LAEB, Kiselev MG (2016) The importance of suppressing spin diffusion effects in the accurate determination of the spatial structure of a flexible molecule by nuclear Overhauser effect spectroscopy. J Mol Struct 1106:373–381
Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ (2007) A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810
Kokryakov VN, Harwig SSL, Panyutich EA, Shevchenko AA, Aleshina GM, Shamova OV, Korneva HA, Lehrer RI (1993) Protegrins—leukocyte antimicrobial peptides that combine features of corticostatic defensins and tachyplesins. FEBS Lett 327:231–236
Kolosova OA, Usachev KS, Aganov AV, Klochkov VV (2016) Antimicrobial peptide protegrins interact with DPC micelles by apolar hydrophobic cluster: structural studies by high-resolution NMR spectroscopy. Bionanosci. doi:10.1007/s12668-016-0218-9
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Langham AA, Khandelia H, Schuster B, Waring AJ, Lehrer RI, Kaznessis YN (2008) Correlation between simulated physicochemical properties and hemolycity of protegrin-like antimicrobial peptides: predicting experimental toxicity. Peptides 29:1085–1093
Lazaridis T, He Y, Prieto L (2013) Membrane interactions and pore formation by the antimicrobial peptide protegrin. Biophys J 104:633–642
Ma Z, Wei DD, Yan P, Zhu X, Shan AS, Bi ZP (2015) Characterization of cell selectivity, physiological stability and endotoxin neutralization capabilities of alpha-helix-based peptide amphiphiles. Biomaterials 52:517–530
Mani R, Tang M, Wu X, Buffy JJ, Waring AJ, Sherman MA, Hong M (2006) Membrane-bound dimer structure of a beta-hairpin antimicrobial peptide from rotational-echo double-resonance solid-state NMR. Biochemistry 45:8341–8349
Matsuzaki K, Murase O, Tokuda H, Funakoshi S, Fujii N, Miyajima K (1994) Orientational and aggregational states of magainin 2 in phospholipid bilayers. Biochemistry 33:3342–3349
Mosca DA, Hurst MA, So W, Viajar BS, Fujii CA, Falla TJ (2000) IB-367, a protegrin peptide with in vitro and in vivo activities against the microflora associated with oral mucositis. Antimicrob Agents Chemother 44(7):1803–1808
Nguyen LT, Haney EF, Vogel HJ (2011) The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol 29:464–472
Niu MF, Chai SM, You XY, Wang WH, Qin CL, Gong Q, Zhang TT, Wan P (2015) Expression of porcine protegrin-1 in Pichia pastoris and its anticancer activity in vitro. Exp Ther Med 9:1075–1079
Panchal RG, Cusack E, Cheley S, Bayley H (1996) Tumor protease-activated, pore-forming toxins from a combinatorial library. Nat Biotechnol 14:852–856
Peschel A, Sahl HG (2006) The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol 4:529–536
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Piotto M, Saudek V, Sklenar V (1992) Gradient-tailored excitation for single-quantum NMR-spectroscopy of aqueous-solutions. J Biomol NMR 2:661–665
Powers JPS, Hancock REW (2003) The relationship between peptide structure and antibacterial activity. Peptides 24:1681–1691
Roumestand C, Louis V, Aumelas A, Grassy G, Calas B, Chavanieu A (1998) Oligomerization of protegrin-1 in the presence of DPC micelles. A proton high-resolution NMR study. FEBS Lett 421:263–267
Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM (2003) The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160:65–73
Shai Y (1999) Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. BBA Biomembr 1462:55–70
Sharma S, Sahoo N, Bhunia A (2016) Antimicrobial peptides and their pore/ion channel properties in neutralization of pathogenic microbes. Curr Top Med Chem 16:46–53
Sklenar V, Piotto M, Leppik R, Saudek V (1993) Gradient-tailored water suppression for H-1-N-15 HSQC experiments optimized to retain full sensitivity. J Magn Reson Ser A 102:241–245
Sokolov Y, Mirzabekov T, Martin DW, Lehrer RI, Kagan BL (1999) Membrane channel formation by antimicrobial protegrins. BBA Biomembr 1420:23–29
Steinberg DA, Hurst MA, Fujii CA, Kung AHC, Ho JF, Cheng FC, Loury DJ, Fiddes JC (1997) Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob Agents Chemother 41:1738–1742
Usachev KS, Filippov AV, Khairutdinov BI, Antzutkin ON, Klochkov VV (2014) NMR structure of the Arctic mutation of the Alzheimer’s A beta(1-40) peptide docked to SDS micelles. J Mol Struct 1076:518–523
Usachev KS, Efimov SV, Kolosova OA, Filippov AV, Klochkov VV (2015a) High-resolution NMR structure of the antimicrobial peptide protegrin-2 in the presence of DPC micelles. J Biomol NMR 61:227–234
Usachev KS, Efimov SV, Kolosova OA, Klochkova EA, Aganov AV, Klochkov VV (2015b) Antimicrobial peptide protegrin-3 adopt an antiparallel dimer in the presence of DPC micelles: a high-resolution NMR study. J Biomol NMR 62:71–79
Vivcharuk V, Kaznessis Y (2010a) Free energy profile of the interaction between a monomer or a dimer of protegrin-1 in a specific binding orientation and a model lipid bilayer. J Phys Chem B 114:2790–2797
Vivcharuk V, Kaznessis YN (2010b) Dimerization of protegrin-1 in different environments. Int J Mol Sci 11:3177–3194
Wade D, Boman A, Wahlin B, Drain CM, Andreu D, Boman HG, Merrifield RB (1990) All-D amino acid-containing channel-forming antibiotic peptides. Proc Natl Acad Sci USA 87:4761–4765
Yang L, Weiss TM, Lehrer RI, Huang HW (2000) Crystallization of antimicrobial pores in membranes: magainin and protegrin. Biophys J 79:2002–2009
Yeaman MR, Yount NY (2003) Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev 55:27–55
Zasloff M (2002) Antimicrobial peptides of multicellular organisms. Nature 415:389–395
Acknowledgments
We thank Dr. Andrey Filippov for the peptide synthesized. The work was performed according to the Russian Government Program of Competitive Growth of Kazan Federal University; by the subsidy allocated to Kazan Federal University for the project part of the state assignment in the sphere of scientific activities; by RFBR, according to the research project No. 16-34-60001 mol_a_dk.
Author information
Authors and Affiliations
Corresponding author
Additional information
Database: Structural data are available in the Protein Data Bank/BioMagResBank databases under the accession numbers 2nc7/26009.
Rights and permissions
About this article
Cite this article
Usachev, K.S., Kolosova, O.A., Klochkova, E.A. et al. Oligomerization of the antimicrobial peptide Protegrin-5 in a membrane-mimicking environment. Structural studies by high-resolution NMR spectroscopy. Eur Biophys J 46, 293–300 (2017). https://doi.org/10.1007/s00249-016-1167-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-016-1167-5