Abstract
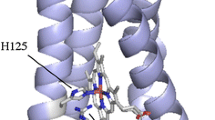
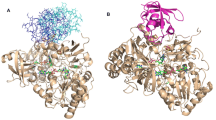
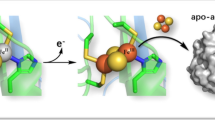
Nitrite reductase (NiR) is a highly stable trimeric protein, which denatures via an intermediate, \({\text{N}}_{3} \overset k \longleftrightarrow {\text{U}}_{3} \xrightarrow{k}{\text{F}} \) (N—native, U—unfolded and F—final). To understand the role of interfacial residues on protein stability, a type-2 copper site ligand, His306, has been mutated to an alanine. The characterization of the native state of the mutated protein highlights that this mutation prevents copper ions from binding to the type-2 site and eliminates catalytic activity. No significant alteration of the geometry of the type-1 site is observed. Study of the thermal denaturation of this His306Ala NiR variant by differential scanning calorimetry shows an endothermic irreversible profile, with maximum heat absorption at T max ≈ 85°C, i.e., 15°C lower than the corresponding value found for wild-type protein. The reduction of the protein thermal stability induced by the His306Ala replacement was also shown by optical spectroscopy. The denaturation pathway of the variant is compatible with the kinetic model \({\text{N}}_{3} \xrightarrow{k}{\text{F}}_{3} , \) where the protein irreversibly passes from the native to the final state. No evidence of subunits’ dissociation has been found within the unfolding process. The results show that the type-2 copper sites, situated at the interface of two monomers, significantly contribute to both the stability and the denaturation mechanism of NiR.





Similar content being viewed by others
References
Adman ET, Godden JW, Turley S (1995) The structure of copper nitrite-reductase from Achromobacter cycloclastes at five pH values, with NO− 2 bound and with type II copper depleted. J Biol Chem 46:27458–27474
Agashe VR, Udgaonkar JB (1995) Thermodynamics of denaturation of barstar: evidence for cold denaturation and evaluation of the interaction with guanidine hydrochloride. Biochemistry 34:3286–3299
Alber T, Matthews BW (1987) Structure and thermal stability of phage T4 lysozyme. Methods Enzymol 154:511–533
Alcaraz LA, Donaire A (2005) Rapid binding of copper(I) to folded aporusticyanin. FEBS Lett 579:5223–5226
Ali MH, Imperiali B (2005) Protein oligomerization: how and why. Bioorg Med Chem 13:5013–5020
Banerjee T, Kishore N (2004) A differential scanning calorimetric study on the irreversible thermal unfolding of concanavalin A. Thermochim Acta 411:195–201
Boulanger MJ, Kukimoto M, Nishiyama M, Horinouchi S, Murphy MEP (2000) Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase. J Biol Chem 275:23957–23964
Brenner AJ, Harris ED (1995) A quantitative test for copper using bicinchoninic acid. Anal Biochem 226:80–84
Chen Y, Mao H, Zhang X, Gong Y, Zhao N (1999) Thermal conformational changes of bovine fibrinogen by differential scanning calorimetry and circular dichroism. Int J Biol Macromol 26:129–134
Edwards RA, Whittaker MM, Whittaker JW, Baker EN, Jameson GB (2001) Removing a hydrogen bond in the dimer interface of Escherichia coli manganese superoxide dismutase alters structure and reactivity. Biochemistry 40:4622–4632
Ellis MJ, Antonyuk SV, Strange RW, Sawers G, Eady RR, Hasnain SS (2004) Observation of an unprecedented Cu bis–His site: crystal structure of the H129V mutant of nitrite reductase. Inorg Chem 43:7591–7593
Freire E, van Osdol WW, Mayorga OL, Sanchez-Ruiz JM (1990) Calorimetrically determined dynamics of complex unfolding transitions in proteins. Annu Rev Biophys Biophys Chem 19:159–188
Godden JW, Turley S, Teller DC, Adman ET, Liu MY, Payne WJ, LeGall J (1991) The 2.3 angstrom X-ray structure of nitrite reductase from Achromobacter cycloclastes. Science 253:438–442
Goodsell DS, Olson AJ (2000) Structural symmetry and protein function. Annu Rev Biophys Biomol Struct 29:105–153
Hough MA, Ellis MJ, Antonyuk SV, Strange RW, Sawers G, Eady RR, Hasnain SS (2005) High resolution structural studies of mutants provide insights into catalysis and electron transfer processes in copper nitrite reductase. J Mol Biol 350:300–309
Jacob C, Holme AL, Fry FH (2004) The sulfinic acid switch in proteins. Org Biomol Chem 2:1953–1956
Kallrot N, Nilsson K, Rasmussen T, Ryde U (2005) Theoretical study of structure of catalytic of copper site in nitrite reductase. Int J Quantum Chem 102:520–541
Koroleva OV, Stepanova EV, Binukov VI, Timofeev VP, Pfeil W (2001) Temperature-induced changes in copper centers and protein conformation of two fungal laccase from Coriolus hirsutus and Coriolus zonatus. Biochim Biophys Acta 1547:397–407
Kurganov BI, Lyubarev AE, Sanchez-Ruiz JM, Shnyrov VL (1997) Analysis of differential scanning calorimetry data for proteins. Criteria of validity of one-step mechanism of irreversible protein denaturation. Biophys Chem 69:125–135
LaCroix LB, Shadle SE, Wang Y, Averill BA, Hedman B, Hodgson KO, Solomon EI (1996) Electronic structure of the perturbed blue copper site in nitrite reductase: spectroscopic properties, bonding, and implications for the entatic/rack state. J Am Chem Soc 118:7755–7768
La Rosa C, Grasso DM, Milardi D, Guzzi R, Sportelli L (1995) Thermodynamics of the thermal unfolding of azurin. J Phys Chem 99:14864–14870
Libby E, Averill BA (1992) Evidence that the type 2 copper centers are the site of nitrite reduction by Achromobacter cycloclastes nitrite reductase. Biochem Biophys Res Commun 187:1529–1535
Lyubarev AE, Kurganov BI, Burlakova AA, Orlov VN (1998) Irreversible thermal denaturation of uridine phosphorylase from Escherichia coli K-12. Biophys Chem 70:247–257
Maithal K, Ravindra G, Nagaraj G, Singh SK, Balaram H, Balaram P (2002) Subunit interface mutation disrupting an aromatic cluster in Plasmodium falciparum triosephosphate isomerase: effect on dimer stability. Protein Eng 15:575–584
McGarvey BR (1967) Electron spin resonance of transition metal complexes. In: Carlin R (ed) Transition metal chemistry, vol 3. M. Dekker, New York, pp 90–201
Murphy MEP, Turley S, Kukimoto M, Nishiyama M, Horinouchi S, Sasaki H, Tanokura M, Adman ET (1995) Structure of Alcaligenes faecalis nitrite reductase and a copper site mutant, M150E, that contains zinc. Biochemistry 34:12107–12117
Murphy MEP, Turley S, Adman ET (1997) Structure of nitrite bound to copper-containing nitrite reductase from Alcaligenes faecalis. J Biol Chem 45:28455–28460
Nishiyama M, Suzuki J, Kukimoto M, Ohnuki T, Horinouchi S, Beppu T (1993) Cloning and characterization of a nitrite reductase gene from Alcaligenes faecalis and its expression in Escherichia coli. J Gen Microbiol 139:725–733
Ramilo CA, Leveque V, Guan Y, Lepock JR, Tainer JA, Nick HS, Silverman DN (1999) Interrupting the hydrogen bond network at the active site of human manganese superoxide dismutase. J Biol Chem 274:27711–27716
Sanchez-Ruiz JM (1992) Theoretical analysis of Lumry–Eyring models in differential scanning calorimetry. Biophys J 61:921–935
Sanchez-Ruiz JM, Lopez-Lacomba JL, Cortijo M, Mateo PL (1988) Differential scanning calorimetry of the irreversible thermal denaturation of thermolysin. Biochemistry 27:1648–1652
Sandberg A, Leckner J, Shi Y, Schwarz FP, Karlsson BG (2002) Effects of metal ligation and oxygen on the reversibility of the thermal denaturation of Pseudomonas aeruginosa azurin. Biochemistry 41:1060–1069
Stirpe A, Guzzi R, Wijma H, Verbeet MPh, Canters GW, Sportelli L (2005) Calorimetric and spectroscopic investigations of the thermal denaturation of wild type nitrite reductase. Biochim Biophys Acta 1752:47–55
Stirpe A, Sportelli L, Guzzi R (2006) A comparative investigation of the thermal unfolding of pseudoazurin in the Cu(II)-holo and apo form. Biopolymers 83:487–497
Suzuki S, Deligeer, Yamaguchi K, Kataoka K, Kobayashi K, Tagawa S, Kohzuma T, Shidara S, Iwasaki H (1997) Spectroscopic characterization and intramolecular electron transfer processes of native and type 2 copper depleted nitrite reductases. J Biol Inor Chem 2:265–274
Thorolfsson M, Ibarra-Molero B, Fojan P, Petersen SB, Sanchez-Ruiz JM, Martinez A (2002) l-phenylalanine binding and domain organization in human phenylalanine hydroxylase: a differential scanning calorimetry study. Biochemistry 41:7573–7585
Tigerstrom A, Schwarz FP, Karlsson BG, Okvist M, Alvarez-Rua C, Maeder D, Robb FT, Sjolin L (2004) Effects of a novel disulfide bond and engineered electrostatic interactions on the thermostability of azurin. Biochemistry 43:12563–12574
Wijma HJ, Boulanger MJ, Molon A, Fittipaldi M, Huber M, Murphy MEP, Verbeet MPh, Canters GW (2003) Reconstitution of the type-1 active site of the H145G/A variants of nitrite reductase by ligand insertion. Biochemistry 42:4075–4083
Zale SE, Klibanov AM (1986) Why does ribonuclease irreversibly inactivate at high temperatures? Biochemistry 25:5432–5444
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Stirpe, A., Sportelli, L., Wijma, H. et al. Thermal stability effects of removing the type-2 copper ligand His306 at the interface of nitrite reductase subunits. Eur Biophys J 36, 805–813 (2007). https://doi.org/10.1007/s00249-007-0151-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-007-0151-5