Abstract
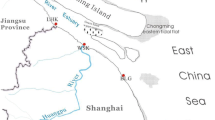
Investigating the environmental influence on the community composition and abundance of denitrifiers in marine sediment ecosystem is essential for understanding of the ecosystem-level controls on the biogeochemical process of denitrification. In the present study, nirK-harboring denitrifying communities in different mud deposit zones of eastern China marginal seas (ECMS) were investigated via clone library analysis. The abundance of three functional genes affiliated with denitrification (narG, nirK, nosZ) was assessed by fluorescent quantitative PCR. The nirK-harboring microbiota were dominated by a few operational taxonomic units (OTUs), which were widely distributed in different sites with each site harboring their unique phylotypes. The mean abundance of nirK was significantly higher than that of narG and nosZ genes, and the abundance of narG was higher than that of nosZ. The inconsistent abundance profile of different functional genes along the process of denitrification might indicate that nitrite reduction occurred independently of denitrification in the mud deposit zones of ECMS, and sedimentary denitrification was accomplished by cooperation of different denitrifying species rather than a single species. Such important information would be missed when targeting only a single denitrifying functional gene. Analysis of correlation between abundance ratios and environmental factors revealed that the response of denitrifiers to environmental factors was not invariable in different mud deposit zones. Our results suggested that a comprehensive analysis of different denitrifying functional genes may gain more information about the dynamics of denitrifying microbiota in marine sediments.







Similar content being viewed by others
References
Gruber N (2008) The marine nitrogen cycle: overview and challenges. In: Douglas GC (ed) Nitrogen in the marine environment, 2nd edn. Academic Press, pp 1–50
Philippot L, Spor A, Hénault C, Bru D, Bizouard F, Jones CM, Sarr A, Maron P-A (2013) Loss in microbial diversity affects nitrogen cycling in soil. ISME J 7:1609–1619. doi:10.1038/ismej.2013.34
Bowen JL, Weisman D, Yasuda M, Jayakumar A, Morrison HG, Ward BB (2015) Marine oxygen-deficient zones harbor depauperate denitrifying communities compared to novel genetic diversity in coastal sediments. Microb Ecol 70:311–321. doi:10.1007/s00248-015-0582-y
Yao P, Zhao B, Bianchi TS, Guo Z, Zhao M, Li D, Pan H, Wang J, Zhang T, Yu Z (2014) Remineralization of sedimentary organic carbon in mud deposits of the Changjiang Estuary and adjacent shelf: implications for carbon preservation and authigenic mineral formation. Cont Shelf Res 91:1–11. doi:10.1016/j.csr.2014.08.010
Knowles R (1982) Denitrification. Microbiol Rev 46:43–70
Zumft WG (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61:533–616
Bru D, Sarr A, Philippot L (2007) Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl Environ Microbiol 73:5971–5974. doi:10.1128/AEM.00643-07
Braker G, Zhou J, Wu L, Devol AH, Tiedje JM (2000) Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl Environ Microbiol 66:2096–2104. doi:10.1128/AEM.66.5.2096-2104.2000
Horn MA, Drake HL, Schramm A (2006) Nitrous oxide reductase genes (nosZ) of denitrifying microbial populations in soil and the earthworm gut are phylogenetically similar. Appl Environ Microbiol 72:1019–1026. doi:10.1128/AEM.72.2.1019-1026.2006
Huang S, Chen C, Yang X, Wu Q, Zhang R (2011) Distribution of typical denitrifying functional genes and diversity of the nirS-encoding bacterial community related to environmental characteristics of river sediments. Biogeosciences 8:3041–3051. doi:10.5194/bg-8-3041-2011
Dandie CE, Wertz S, Leclair CL, Goyer C, Burton DL, Patten CL, Zebarth BJ, Trevors JT (2011) Abundance, diversity and functional gene expression of denitrifier communities in adjacent riparian and agricultural zones. FEMS Microbiol Ecol 77:69–82. doi:10.1111/j.1574-6941.2011.01084.x
Chon K, Cho J (2015) Abundance and expression of denitrifying genes (narG, nirS, norB, and nosZ) in sediments of wastewater stabilizing constructed wetlands. Environ Eng Res 20:51–57. doi:10.4491/eer.2014.069
Bárta J, Melichová T, Vaněk D, Picek T, Šantrůčková H (2010) Effect of pH and dissolved organic matter on the abundance of nirK and nirS denitrifiers in spruce forest soil. Biogeochemistry 101:123–132. doi:10.1007/s10533-010-9430-9
Crutzen PJ (1979) The role of NO and NO2 in the chemistry of the troposphere and stratosphere. Annu Rev Earth Planet Sci 7:443–472
Henry S, Bru D, Stres B, Hallet S, Philippot L (2006) Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl Environ Microbiol 72:5181–5189. doi:10.1128/AEM.00231-06
Smith CJ, Nedwell DB, Dong LF, Osborn AM (2007) Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl Environ Microbiol 73:3612–3622. doi:10.1128/AEM.02894-06
Liu J, Xu K, Aea L, Milliman J, Velozzi D, Xiao S, Yang Z (2007) Flux and fate of Yangtze River sediment delivered to the East China Sea. Geomorphology 85:208–224. doi:10.1016/j.geomorph.2006.03.023
Hu L, Shi X, Guo Z, Wang H, Yang Z (2013) Sources, dispersal and preservation of sedimentary organic matter in the Yellow Sea: the importance of depositional hydrodynamic forcing. Mar Geol 335:52–63. doi:10.1016/j.margeo.2012.10.008
Yu S, Yao P, Liu J, Zhao B, Zhang G, Zhao M, Yu Z, Zhang X-H (2016) Diversity, abundance, and niche differentiation of ammonia-oxidizing prokaryotes in mud deposits of the eastern China marginal seas. Front Microbiol 7:137. doi:10.3389/fmicb.2016.00137
Magalhães C, Bano N, Wiebe W, Bordalo A, Hollibaugh J (2008) Dynamics of nitrous oxide reductase genes (nosZ) in intertidal rocky biofilms and sediments of the Douro River Estuary (Portugal), and their relation to N-biogeochemistry. Microb Ecol 55:259–269. doi:10.1007/s00248-007-9273-7
Chon K, Chang J-S, Lee E, Lee J, Ryu J, Cho J (2011) Abundance of denitrifying genes coding for nitrate (narG), nitrite (nirS), and nitrous oxide (nosZ) reductases in estuarine versus wastewater effluent-fed constructed wetlands. Ecol Eng 37:64–69. doi:10.1016/j.ecoleng.2009.04.005
Wu L, Osmond DL, Graves AK, Burchell MR, Duckworth OW (2012) Relationships between nitrogen transformation rates and gene abundance in a riparian buffer soil. Environ Manag 50:861–874. doi:10.1007/s00267-012-9929-z
Jayakumar DA, Francis CA, Naqvi SWA, Ward BB (2004) Diversity of nitrite reductase genes (nirS) in the denitrifying water column of the coastal Arabian Sea. Aquat Microb Ecol 34:69–78. doi:10.3354/ame034069
Ward B, Devol A, Rich J, Chang B, Bulow S, Naik H, Pratihary A, Jayakumar A (2009) Denitrification as the dominant nitrogen loss process in the Arabian Sea. Nature 461:78–81. doi:10.1038/nature08276
Li M, Hong Y, Cao H, Klotz MG, Gu JD (2013) Diversity, abundance, and distribution of NO-forming nitrite reductase-encoding genes in deep-sea subsurface sediments of the South China Sea. Geobiology 11:170–179. doi:10.1111/gbi.12020
Vilar-Sanz A, Puig S, García-Lledó A, Trias R, Balaguer MD, Colprim J, Bañeras L (2013) Denitrifying bacterial communities affect current production and nitrous oxide accumulation in a microbial fuel cell. PLoS ONE 8:e63460. doi:10.1371/journal.pone.0063460
Hu L, Shi X, Yu Z, Lin T, Wang H, Ma D, Guo Z, Yang Z (2012) Distribution of sedimentary organic matter in estuarine–inner shelf regions of the East China Sea: implications for hydrodynamic forces and anthropogenic impact. Mar Chem 142:29–40. doi:10.1016/j.marchem.2012.08.004
Chen Z, Saito Y, Kanai Y, Wei T, Li L, Yao H, Wang Z (2004) Low concentration of heavy metals in the Yangtze estuarine sediments, China: a diluting setting. Estuar Coast Shelf Sci 60:91–100. doi:10.1016/j.ecss.2003.11.021
Huh C-A, Su C-C (1999) Sedimentation dynamics in the East China Sea elucidated from 210Pb, 137Cs and 239,240 Pu. Mar Geol 160:183–196. doi:10.1016/S0025-3227(99)00020-1
Liu J, Zhu R, Li G (2003) Rock magnetic properties of the fine-grained sediment on the outer shelf of the East China Sea: implication for provenance. Mar Geol 193:195–206. doi:10.1016/S0025-3227(02)00660-6
Yang SY, Jung HS, Lim DI, Li CX (2003) A review on the provenance discrimination of sediments in the Yellow Sea. Earth-Sci Rev 63:93–120. doi:10.1016/S0012-8252(03)00033-3
Lim D, Choi J, Jung H, Rho K, Ahn K (2007) Recent sediment accumulation and origin of shelf mud deposits in the Yellow and East China Seas. Prog Oceanogr 73:145–159. doi:10.1016/j.pocean.2007.02.004
Throbäck IN, Enwall K, Jarvis Å, Hallin S (2004) Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Ecol 49:401–417. doi:10.1016/j.femsec.2004.04.011
Hallin S, Lindgren PE (1999) PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl Environ Microbiol 65:1652–1657
Hallin S, Jones CM, Schloter M, Philippot L (2009) Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J 3:597–605. doi:10.1038/ismej.2008.128
Hill AR, Cardaci M (2004) Denitrification and organic carbon availability in riparian wetland soils and subsurface sediments. Soil Sci Soc Am J 68:320–325. doi:10.2136/sssaj2004.3200
Liu J, Hou H, Sheng R, Chen Z, Zhu Y, Qin H, Wei W (2012) Denitrifying communities differentially respond to flooding drying cycles in paddy soils. Appl Soil Ecol 62:155–162. doi:10.1016/j.apsoil.2012.06.010
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi:10.1093/molbev/msr121
Farnelid H, Bentzon-Tilia M, Andersson AF, Bertilsson S, Jost G, Labrenz M, Jürgens K, Riemann L (2013) Active nitrogen-fixing heterotrophic bacteria at and below the chemocline of the central Baltic Sea. ISME J 7:1413–1423. doi:10.1038/ismej.2013.26
Larkin MA, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506. doi:10.1128/AEM.71.3.1501-1506.2005
Ter Braak CJ, Smilauer P (2002) CANOCO reference manual and CanoDraw for Windows user’s guide: software for canonical community ordination (version 4.5). www. canoco. com
Dang H, Chen R, Wang L, Guo L, Chen P, Tang Z, Tian F, Li S, Klotz MG (2010) Environmental factors shape sediment anammox bacterial communities in hypernutrified Jiaozhou Bay, China. Appl Environ Microbiol 76:7036–7047. doi:10.1128/AEM.01264-10
Dang H, Luan X-W, Chen R, Zhang X, Guo L, Klotz MG (2010) Diversity, abundance and distribution of amoA-encoding archaea in deep-sea methane seep sediments of the Okhotsk Sea. FEMS Microbiol Ecol 72:370–385. doi:10.1111/j.1574-6941.2010.00870.x
Lozupone CA, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. doi:10.1128/AEM.01996-06
Rösch C, Mergel A, Bothe H (2002) Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl Environ Microbiol 68:3818–3829. doi:10.1128/AEM.68.8.3818-3829.2002
Guo G-X, Deng H, Qiao M, Yao H-Y, Zhu Y-G (2013) Effect of long-term wastewater irrigation on potential denitrification and denitrifying communities in soils at the watershed scale. Environ Sci Technol 47:3105–3113. doi:10.1021/es304714a
Hartmann AA, Barnard RL, Marhan S, Niklaus PA (2013) Effects of drought and N-fertilization on N cycling in two grassland soils. Oecologia 171:705–717. doi:10.1007/s00442-012-2578-3
Yoshida M, Ishii S, Otsuka S, Senoo K (2010) nirK-harboring denitrifiers are more responsive to denitrification-inducing conditions in rice paddy soil than nirS-harboring bacteria. Microbes Environ 25:45–48. doi:10.1264/jsme2.ME09160
Mosier AC, Francis CA (2010) Denitrifier abundance and activity across the San Francisco Bay estuary. Environ Microbiol Rep 2:667–676. doi:10.1111/j.1758-2229.2010.00156.x
Li J, Wei G, Wang N, Gao Z (2014) Diversity and distribution of nirK-harboring denitrifying bacteria in the water column in the Yellow River Estuary. Microbes Environ 29:107–110. doi:10.1264/jsme2.ME13111
Rich J, Heichen R, Bottomley P, Cromack K, Myrold D (2003) Community composition and functioning of denitrifying bacteria from adjacent meadow and forest soils. Appl Environ Microbiol 69:5974–5982. doi:10.1128/AEM.69.10.5974-5982.2003
Dang H, Wang C, Li J, Li T, Tian F, Jin W, Ding Y, Zhang Z (2009) Diversity and distribution of sediment nirS-encoding bacterial assemblages in response to environmental gradients in the eutrophied Jiaozhou Bay, China. Microb Ecol 58:161–169. doi:10.1007/s00248-008-9469-5
Zheng Y, Hou L, Liu M, Gao J, Yin G, Li X, Deng F, Lin X, Jiang X, Chen F (2015) Diversity, abundance, and distribution of nirS-harboring denitrifiers in intertidal sediments of the Yangtze Estuary. Microb Ecol 70:30–40. doi:10.1007/s00248-015-0567-x
Katsuyama C, Kondo N, Suwa Y, Yamagishi T, Itoh M, Ohte N, Kimura H, Nagaosa K, Kato K (2008) Denitrification activity and relevant bacteria revealed by nitrite reductase gene fragments in soil of temperate mixed forest. Microbes Environ 23:337–345. doi:10.1264/jsme2.ME08541
Zhang X, Agogué H, Dupuy C et al (2014) Relative abundance of ammonia oxidizers, denitrifiers, and anammox bacteria in sediments of hyper-nutrified estuarine tidal flats and in relation to environmental conditions. [J] CLEAN–Soil, Air, Water 42(6):815–823. doi:10.1002/clen.201300013
Chen Z, Liu J, Wu M, Xie X, Wu J, Wei W (2012) Differentiated response of denitrifying communities to fertilization regime in paddy soil. Microb Ecol 63:446–459. doi:10.1007/s00248-011-9909-5
Voss M, Bange HW, Dippner JW, Middelburg JJ, Montoya JP, Ward B (2013) The marine nitrogen cycle: recent discoveries, uncertainties and the potential relevance of climate change. Philos Trans R Soc B 368. doi: 10.1098/rstb.2013.0121
Orellana LH, Rodriguez-R LM, Higgins S, Chee-Sanford JC, Sanford RA, Ritalahti KM, Löffler FE, Konstantinidis KT (2014) Detecting nitrous oxide reductase (nosZ) genes in soil metagenomes: method development and implications for the nitrogen cycle. MBio 5:e01193–14. doi:10.1128/mBio.01193-14
Jetten MS (2008) The microbial nitrogen cycle. Environ Microbiol 10:2903–2909. doi:10.1111/j.1462-2920.2008.01786.x
Acknowledgements
We are grateful to the captain and crew of the R/V Dong Fang Hong 2 for their assistance during the cruise. We also appreciate all colleagues who contributed to this study. This work was supported by the National Natural Science Foundation of China through grants 41521064, 41476112, and 41506154.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Additional information
Minghong Gao and Jiwen Liu contributed equally to this work.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 327 kb)
Rights and permissions
About this article
Cite this article
Gao, M., Liu, J., Qiao, Y. et al. Diversity and Abundance of the Denitrifying Microbiota in the Sediment of Eastern China Marginal Seas and the Impact of Environmental Factors. Microb Ecol 73, 602–615 (2017). https://doi.org/10.1007/s00248-016-0906-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0906-6