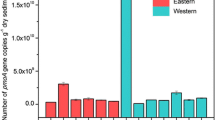
Abstract
Nitrite-oxidizing bacteria (NOB) are chemolithoautotrophs that catalyze the oxidation of nitrite to nitrate, which is the second step of aerobic nitrification. In marine ecosystems, Nitrospina is assumed to be a major contributor to nitrification. To date, two strains of Nitrospina have been isolated from marine environments. Despite their ecological relevance, their ecophysiology and environmental distribution are understudied owing to fastidious cultivation techniques and the lack of a sufficient functional gene marker. To estimate the abundance, diversity, and distribution of Nitrospina in various marine sediments, we used nxrA, which encodes the alpha subunit of nitrite oxidoreductase, as a functional and phylogenetic marker. We observed that Nitrospina diversity in polar sediments was significantly lower than that of non-polar samples. Moreover, nxrA-like sequences revealed an unexpected diversity of Nitrospina, with approximately 41,000 different sequences based on a 95% similarity cutoff from six marine sediments. We detected nxrA gene copy numbers of up to 3.57 × 104 per gram of marine sediment sample. The results of this study provide insight into the distribution and diversity of Nitrospina, which is fundamentally important for understanding their contribution to the nitrogen cycle in marine sediments.





Similar content being viewed by others
References
Jetten MS, Strous M, van de Pas-Schoonen KT, Schalk J, van Dongen UG, van de Graaf AA, Logemann S, Muyzer G, van Loosdrecht MC, Kuenen JG (1998) The anaerobic oxidation of ammonium. FEMS Microbiol Rev 22:421–437
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops HP, Wagner M (2000) Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol 66:5368–5382
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102:14683–14688. doi:10.1073/pnas.0506625102
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809. doi:10.1038/nature04983
Park SJ, Park BJ, Rhee SK (2008) Comparative analysis of archaeal 16S rRNA and amoA genes to estimate the abundance and diversity of ammonia-oxidizing archaea in marine sediments. Extremophiles 12:605–615. doi:10.1007/s00792-008-0165-7
Junier P, Molina V, Dorador C, Hadas O, Kim OS, Junier T, Witzel JP, Imhoff JF (2010) Phylogenetic and functional marker genes to study ammonia-oxidizing microorganisms (AOM) in the environment. Appl Microbiol Biotechnol 85:425–440. doi:10.1007/s00253-009-2228-9
Ish-Am O, Kristensen DM, Ruppin E (2015) Evolutionary conservation of bacterial essential metabolic genes across all bacterial culture media. PLoS ONE 10:e0123785. doi:10.1371/journal.pone.0123785
Eisen JA (1995) The RecA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of RecAs and 16S rRNAs from the same species. J Mol Evol 41:1105–1123
He Z, Gentry TJ, Schadt CW, Wu L, Liebich J, Chong SC, Huang Z, Wu W, Gu B, Jardine P, Criddle C, Zhou J (2007) GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J 1:67–77. doi:10.1038/ismej.2007.2
Case RJ, Boucher Y, Dahllöf I, Holmström C, Doolittle WF, Kjelleberg S (2007) Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol 73:278–288. doi:10.1128/AEM.01177-06
Herbert RA (1999) Nitrogen cycling in coastal marine ecosystems. FEMS Microbiol Rev 23:563–590
Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW (2013) Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. doi:10.1038/nature12375
Spieck E, Bock E (2005) The lithoautotrophic nitrite-oxidizing bacteria. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM (eds) Bergey’s Manual® of systematic bacteriology: volume two: the Proteobacteria, part A Introductory essays. Springer US, Boston, pp 149–153
Garrity GM, Holt JG, Spieck E, Bock E, Johnson DB, Spring S, Schleifer K-H, Maki JS (2001) Phylum BVIII. Nitrospirae phy. nov. In: Boone DR, Castenholz RW, Garrity GM (eds) Bergey’s Manual® of systematic bacteriology: volume one: the Archaea and the deeply branching and phototrophic bacteria. Springer New York, New York, pp 451–464
Sorokin DY, Lucker S, Vejmelkova D, Kostrikina NA, Kleerebezem R, Rijpstra WI, Damste JS, Le Paslier D, Muyzer G, Wagner M, van Loosdrecht MC, Daims H (2012) Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J 6:2245–2256. doi:10.1038/ismej.2012.70
Yamanaka T, Fukumori Y (1988) The nitrite oxidizing system of Nitrobacter winogradskyi. FEMS Microbiol Rev 4:259–270
Watson SW, Waterbury JB Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Arch Mikrobiol 77:203–230. doi: 10.1007/bf00408114
van Kessel MA, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJ, Kartal B, Jetten MS, Lucker S (2015) Complete nitrification by a single microorganism. Nature 528:555–559. doi:10.1038/nature16459
Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M (2015) Complete nitrification by Nitrospira bacteria. Nature 528:504–509. doi:10.1038/nature16461
Koch H, Galushko A, Albertsen M, Schintlmeister A, Gruber-Dorninger C, Lucker S, Pelletier E, Le Paslier D, Spieck E, Richter A, Nielsen PH, Wagner M, Daims H (2014) Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science 345:1052–1054. doi:10.1126/science.1256985
Haaijer SC, Ji K, van Niftrik L, Hoischen A, Speth D, Jetten MS, Damste JS, Op den Camp HJ (2013) A novel marine nitrite-oxidizing Nitrospira species from Dutch coastal North Sea water. Front Microbiol 4:60. doi:10.3389/fmicb.2013.00060
Spieck E, Keuter S, Wenzel T, Bock E, Ludwig W (2014) Characterization of a new marine nitrite oxidizing bacterium, Nitrospina watsonii sp. nov., a member of the newly proposed phylum “Nitrospinae”. Syst Appl Microbiol 37:170–176. doi:10.1016/j.syapm.2013.12.005
Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF (2007) Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol 9:1162–1175. doi:10.1111/j.1462-2920.2007.01239.x
Santoro AE, Casciotti KL, Francis CA (2010) Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ Microbiol 12:1989–2006. doi:10.1111/j.1462-2920.2010.02205.x
Park BJ, Park SJ, Yoon DN, Schouten S, Sinninghe Damste JS, Rhee SK (2010) Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria. Appl Environ Microbiol 76:7575–7587. doi:10.1128/AEM.01478-10
Beman JM, Leilei Shih J, Popp BN (2013) Nitrite oxidation in the upper water column and oxygen minimum zone of the eastern tropical North Pacific Ocean. ISME J 7:2192–2205. doi:10.1038/ismej.2013.96
Lüke C, Speth DR, Kox MA, Villanueva L, Jetten MS (2016) Metagenomic analysis of nitrogen and methane cycling in the Arabian Sea oxygen minimum zone. PeerJ 4:e1924. doi:10.7717/peerj.1924
Lücker S, Nowka B, Rattei T, Spieck E, Daims H (2013) The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol 4:27. doi:10.3389/fmicb.2013.00027
Ngugi DK, Blom J, Stepanauskas R, Stingl U (2016) Diversification and niche adaptations of Nitrospina-like bacteria in the polyextreme interfaces of Red Sea brines. ISME J 10:1383–1399. doi:10.1038/ismej.2015.214
Levipan HA, Molina V, Fernandez C (2014) Nitrospina-like bacteria are the main drivers of nitrite oxidation in the seasonal upwelling area of the Eastern South Pacific (Central Chile approximately 36 degrees S). Environ Microbiol Rep 6:565–573
Regan JM, Harrington GW, Baribeau H, De Leon R, Noguera DR (2003) Diversity of nitrifying bacteria in full-scale chloraminated distribution systems. Water Res 37:197–205
Poly F, Wertz S, Brothier E, Degrange V (2008) First exploration of Nitrobacter diversity in soils by a PCR cloning-sequencing approach targeting functional gene nxrA. FEMS Microbiol Ecol 63:132–140. doi:10.1111/j.1574-6941.2007.00404.x
Vanparys B, Spieck E, Heylen K, Wittebolle L, Geets J, Boon N, De Vos P (2007) The phylogeny of the genus Nitrobacter based on comparative rep-PCR, 16S rRNA and nitrite oxidoreductase gene sequence analysis. Syst Appl Microbiol 30:297–308. doi:10.1016/j.syapm.2006.11.006
Wertz S, Poly F, Le Roux X, Degrange V (2008) Development and application of a PCR-denaturing gradient gel electrophoresis tool to study the diversity of Nitrobacter-like nxrA sequences in soil. FEMS Microbiol Ecol 63:261–271. doi:10.1111/j.1574-6941.2007.00416.x
Pester M, Maixner F, Berry D, Rattei T, Koch H, Lucker S, Nowka B, Richter A, Spieck E, Lebedeva E, Loy A, Wagner M, Daims H (2014) NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ Microbiol 16:3055–3071. doi:10.1111/1462-2920.12300
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. doi:10.1371/journal.pone.0027310
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi:10.2307/2408678
Ramette A (2007) Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62:142–160. doi:10.1111/j.1574-6941.2007.00375.x
Park SJ, Park BJ, Jung MY, Kim SJ, Chae JC, Roh Y, Forwick M, Yoon HI, Rhee SK (2011) Influence of deglaciation on microbial communities in marine sediments off the coast of Svalbard, Arctic Circle. Microb Ecol 62:537–548. doi:10.1007/s00248-011-9860-5
Jongman RHG, ter Braak CJF, OFR VT (1995) Data analysis in community and landscape ecology. Cambridge University Press, Cambridge
Mann HB, Whitney DR (1947) On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat 18:50–60
Füssel J, Lam P, Lavik G, Jensen MM, Holtappels M, Gunter M, Kuypers MM (2012) Nitrite oxidation in the Namibian oxygen minimum zone. ISME J 6:1200–1209. doi:10.1038/ismej.2011.178
Choi H, Koh HW, Kim H, Chae JC, Park SJ (2016) Microbial community composition in the marine sediments of Jeju Island: next-generation sequencing surveys. J Microbiol Biotechnol 26:883–890. doi:10.4014/jmb.1512.12036
Mao DP, Zhou Q, Chen CY, Quan ZX (2012) Coverage evaluation of universal bacterial primers using the metagenomic datasets. BMC Microbiol 12:66. doi:10.1186/1471-2180-12-66
Poretsky R, Rodriguez RL, Luo C, Tsementzi D, Konstantinidis KT (2014) Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One 9:e93827. doi:10.1371/journal.pone.0093827
Boucher Y, Douady CJ, Papke RT, Walsh DA, Boudreau ME, Nesbo CL, Case RJ, Doolittle WF (2003) Lateral gene transfer and the origins of prokaryotic groups. Annu Rev Genet 37:283–328. doi:10.1146/annurev.genet.37.050503.084247
Attard E, Poly F, Commeaux C, Laurent F, Terada A, Smets BF, Recous S, Roux XL (2010) Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ Microbiol 12:315–326. doi:10.1111/j.1462-2920.2009.02070.x
Gregory LG, Bond PL, Richardson DJ, Spiro S (2003) Characterization of a nitrate-respiring bacterial community using the nitrate reductase gene (narG) as a functional marker. Microbiology 149:229–237. doi:10.1099/mic.0.25849-0
Junge K, Eicken H, Deming JW (2004) Bacterial activity at −2 to −20 degrees C in Arctic wintertime sea ice. Appl Environ Microbiol 70:550–557
Alonso-Sáez L, Galand PE, Casamayor EO, Pedrós-Alió C, Bertilsson S (2010) High bicarbonate assimilation in the dark by Arctic bacteria. ISME J 4:1581–1590. doi:10.1038/ismej.2010.69
Alawi M, Lipski A, Sanders T, Pfeiffer EM, Spieck E (2007) Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J 1:256–264
Alawi M, Off S, Kaya M, Spieck E (2009) Temperature influences the population structure of nitrite-oxidizing bacteria in activated sludge. Environ Microbiol Rep 1:184–190. doi:10.1111/j.1758-2229.2009.00029.x
Christner BC, Priscu JC, Achberger AM, Barbante C, Carter SP, Christianson K, Michaud AB, Mikucki JA, Mitchell AC, Skidmore ML, Vick-Majors TJ, Team WS (2014) A microbial ecosystem beneath the West Antarctic ice sheet. Nature 512:310–313. doi:10.1038/nature13667
Kim JG, Park SJ, Quan ZX, Jung MY, Cha IT, Kim SJ, Kim KH, Yang EJ, Kim YN, Lee SH, Rhee SK (2014) Unveiling abundance and distribution of planktonic Bacteria and Archaea in a polynya in Amundsen Sea, Antarctica. Environ Microbiol 16:1566–1578. doi:10.1111/1462-2920.12287
Sun MY, Dafforn KA, Johnston EL, Brown MV (2013) Core sediment bacteria drive community response to anthropogenic contamination over multiple environmental gradients. Environ Microbiol 15:2517–2531. doi:10.1111/1462-2920.12133
Hernández DL, Hobbie SE (2010) The effects of substrate composition, quantity, and diversity on microbial activity. Plant Soil 335:397–411. doi:10.1007/s11104-010-0428-9
Gillan DC, Danis B, Pernet P, Joly G, Dubois P (2005) Structure of sediment-associated microbial communities along a heavy-metal contamination gradient in the marine environment. Appl Environ Microbiol 71:679–690. doi:10.1128/AEM.71.2.679-690.2005
Koch H, Lucker S, Albertsen M, Kitzinger K, Herbold C, Spieck E, Nielsen PH, Wagner M, Daims H (2015) Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc Natl Acad Sci U S A 112:11371–11376. doi:10.1073/pnas.1506533112
Daims H, Maixner F, Lucker S, Stoecker K, Hace K, Wagner M (2006) Ecophysiology and niche differentiation of Nitrospira-like bacteria, the key nitrite oxidizers in wastewater treatment plants. Water Sci Technol 54:21–27
Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M, Daims H (2006) Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol 8:1487–1495. doi:10.1111/j.1462-2920.2006.01033.x
Gruber-Dorninger C, Pester M, Kitzinger K, Savio DF, Loy A, Rattei T, Wagner M, Daims H (2015) Functionally relevant diversity of closely related Nitrospira in activated sludge. ISME J 9:643–655. doi:10.1038/ismej.2014.156
Fujitani H, Aoi Y, Tsuneda S (2013) Selective enrichment of two different types of Nitrospira-like nitrite-oxidizing bacteria from a wastewater treatment plant. Microbes Environ 28:236–243
Starkenburg SR, Chain PS, Sayavedra-Soto LA, Hauser L, Land ML, Larimer FW, Malfatti SA, Klotz MG, Bottomley PJ, Arp DJ, Hickey WJ (2006) Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl Environ Microbiol 72:2050–2063. doi:10.1128/AEM.72.3.2050-2063.2006
Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, Rattei T, Damste JS, Spieck E, Le Paslier D, Daims H (2010) A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci U S A 107:13479–13484. doi:10.1073/pnas.1003860107
Grein F, Ramos AR, Venceslau SS, Pereira IA (2013) Unifying concepts in anaerobic respiration: insights from dissimilatory sulfur metabolism. Biochim Biophys Acta 1827:145–160. doi:10.1016/j.bbabio.2012.09.001
Muller JA, DasSarma S (2005) Genomic analysis of anaerobic respiration in the archaeon Halobacterium sp. strain NRC-1: dimethyl sulfoxide and trimethylamine N-oxide as terminal electron acceptors. J Bacteriol 187:1659–1667. doi:10.1128/JB.187.5.1659-1667.2005
Acknowledgments
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (no. 2015R1C1A1A01053750) and the C1 Gas Refinery Program through the NRF funded by the Ministry of Science, ICT & Future Planning (no. 2015M3D3A1A01064881). We thank the editor and two anonymous reviewers for their constructive and scientific comments, which helped us to improve the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 751 kb)
Rights and permissions
About this article
Cite this article
Rani, S., Koh, HW., Rhee, SK. et al. Detection and Diversity of the Nitrite Oxidoreductase Alpha Subunit (nxrA) Gene of Nitrospina in Marine Sediments. Microb Ecol 73, 111–122 (2017). https://doi.org/10.1007/s00248-016-0897-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0897-3