Abstract
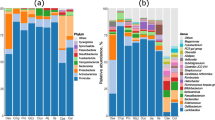
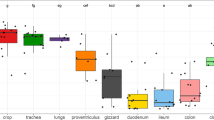
Gastrointestinal microbiota is increasingly recognized as an important component of individual health, and therefore, our ability to quantify its diversity accurately is central for exploring different ways to improve health. Non-invasive sampling methods, such as cloaca swabs, are often used to measure gastrointestinal microbiota diversity within an individual. However, few studies have addressed to what degree differences exist in microbial community composition along the gastrointestinal tract, and measures obtained from the cloaca may not actually represent the diversity present elsewhere in the gastrointestinal tract. In this study, we systematically characterized the gastrointestinal microbial community of the critically endangered Attwater’s Prairie chicken (Tympanuchus cupido attwateri) by opportunistically sampling four different locations (ileum, cecum, large intestine, and cloaca) along the gastrointestinal tract of eight individuals. Spatial variation of microbial community was observed at different sampling locations within the gastrointestinal tract. The cecum harbored the most diverse and significantly different microbiota from the other locations, while the microbial α- and β-diversities were similar in the ileum, large intestine, and cloaca. The results of this study provide evidence that microbiota diversity can differ depending on sampling location and metric used to quantify diversity. As shown here, non-invasive cloacal sampling strategies may reflect microbiota diversity elsewhere in the gastrointestinal tract, yet caution is warranted when making generalizations in terms of the microbiota diversity correlations when samples are obtained from a single location within the gastrointestinal tract.






Similar content being viewed by others
References
Clemente Jose C, Ursell Luke K, Parfrey Laura W, Knight R (2012) The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. doi:10.1016/j.cell.2012.01.035
Clavel T, Desmarchelier C, Haller D, Gérard P, Rohn S, Lepage P, Daniel H (2014) Intestinal microbiota in metabolic diseases. Gut Microbes 5:544–551. doi:10.4161/gmic.29331
Dethlefsen L, McFall-Ngai M, Relman D (2007) An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449:811–818
Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE (2006) Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359. doi:10.1126/science.1124234
Ritchie LE, Steiner JM, Suchodolski JS (2008) Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis. FEMS Microbiol Ecol 66:590–598. doi:10.1111/j.1574-6941.2008.00609.x
Macpherson A, Harris N (2004) Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 4:478–485
Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, Taylor TD, Noguchi H, Mori H, Ogura Y, Ehrlich DS, Itoh K, Takagi T, Sakaki Y, Hayashi T, Hattori M (2007) Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res 14:169–181. doi:10.1093/dnares/dsm018
Lu J, Idris U, Harmon B, Hofacre C, Maurer JJ, Lee MD (2003) Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl Environ Microbiol 69:6816–6824. doi:10.1128/aem.69.11.6816-6824.2003
Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS (2013) Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339:1084–1088
Wang M, Ahrné S, Jeppsson B, Molin G (2005) Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol 54:219–231. doi:10.1016/j.femsec.2005.03.012
Yeoman CJ, Chia N, Jeraldo P, Sipos M, Goldenfeld ND, White BA (2012) The microbiome of the chicken gastrointestinal tract. Anim Health Res Rev 13:89–99
van der Wielen PWJJ, Keuzenkamp DA, Lipman LJA, van Knapen F, Biesterveld S (2002) Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb Ecol 44:286–293. doi:10.1007/s00248-002-2015-y
Su H, McKelvey J, Rollins D, Zhang M, Brightsmith DJ, Derr J, Zhang S (2014) Cultivable bacterial microbiota of northern bobwhite (Colinus virginianus): a new reservoir of antimicrobial resistance? PLoS One 9:e99826. doi:10.1371/journal.pone.0099826
Pryde SE, Richardson AJ, Stewart CS, Flint HJ (1999) Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl Environ Microbiol 65:5372–5377
McLelland J (1989) Anatomy of the avian cecum. J Exp Zool 252:2–9. doi:10.1002/jez.1402520503
Gelis S (2006) Evaluating and treating the gastrointestinal system. Spix Publishing, Inc., Palm Beach
Barnes EM, Mead GC, Barnum DA, Harry EG (1972) The intestinal flora of the chicken in the period 2 to 6 weeks of age, with particular reference to the anaerobic bacteria. Br Poult Sci 13:311–326
Salanitro JP, Fairchilds IG, Zgornicki YD (1974) Isolation, culture characteristics, and identification of anaerobic bacteria from the chicken cecum. Appl Microbiol 27:678–687
Ruiz-RodrÍGuez M, Lucas FS, Heeb P, Soler JJ (2009) Differences in intestinal microbiota between avian brood parasites and their hosts. Biol J Linn Soc 96:406–414. doi:10.1111/j.1095-8312.2008.01127.x
Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB (2012) Animal behavior and the microbiome. Science 338:198–199
Ruiz-Rodríguez M, Soler JJ, Lucas FS, Heeb P, José Palacios M, Martín-Gálvez D, De Neve L, Pérez-Contreras T, Martínez JG, Soler M (2009) Bacterial diversity at the cloaca relates to an immune response in magpie Pica pica and to body condition of great spotted cuckoo Clamator glandarius nestlings. J Avian Biol 40:42–48. doi:10.1111/j.1600-048X.2008.04471.x
van Dongen WF, White J, Brandl H, Moodley Y, Merkling T, Leclaire S, Blanchard P, Danchin E, Hatch S, Wagner R (2013) Age-related differences in the cloacal microbiota of a wild bird species. BMC Ecol 13:11
Leticia Mirón AM, Rocha-Ramírez V, Belda-Ferre P, Cabrera-Rubio R, Folch-Mallol J, Cardénas-Vázquez R, DeLuna A, Lilia Hernández A, Maya-Elizarrarás E, and Schondube JE (2014) Gut bacterial diversity of the house sparrow (Passer domesticus) inferred by 16S rRNA sequence analysis. Metagenomics 3
Morrow ME, Rossignol TA, Silvy NJ (2004) Federal listing of prairie grouse: lessons from the Attwater’s prairie-chicken. Wildl Soc Bull 32:112–118. doi:10.2193/0091-7648(2004)32[112:FLOPGL]2.0.CO;2
Caporaso J, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci 108:4516–4522
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi:10.1128/aem.01541-09
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi:10.1093/bioinformatics/btr381
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi:10.1128/aem.00062-07
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Stat 11:265–270
Chazdon RL, Colwell RK, Denslow JS, Guariguata MR (1998) Statistical methods for estimating species richness of woody regeneration in primary and secondary rain forests of Northeastern Costa Rica. In: Dallmeier F, Comiskey JA (eds) Forest biodiversity research, monitoring and modeling: conceptual background and old world case studies. Parthenon Publishing, Paris, France., pp 285–309
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi:10.1128/aem.71.12.8228-8235.2005
Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143. doi:10.1111/j.1442-9993.1993.tb00438.x
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B (Stat Method) 57:289–300. doi:10.2307/2346101
Choi JH, Kim GB, Cha CJ (2014) Spatial heterogeneity and stability of bacterial community in the gastrointestinal tracts of broiler chickens. Poult Sci 93:1942–1950. doi:10.3382/ps.2014-03974
Rehman HU, Vahjen W, Awad WA, Zentek J (2007) Indigenous bacteria and bacterial metabolic products in the gastrointestinal tract of broiler chickens. Arch Anim Nutr 61:319–335. doi:10.1080/17450390701556817
Dillon RJ, Vennard CT, Buckling A, Charnley AK (2005) Diversity of locust gut bacteria protects against pathogen invasion. Ecol Lett 8:1291–1298. doi:10.1111/j.1461-0248.2005.00828.x
Girvan MS, Campbell CD, Killham K, Prosser JI, Glover LA (2005) Bacterial diversity promotes community stability and functional resilience after perturbation. Environ Microbiol 7:301–313. doi:10.1111/j.1462-2920.2005.00695.x
Ussery H (2011) The small-scale poultry flock: an all-natural approach to raising chickens and other fowl for home and market growers. Chelsea Green Publishing, Chelsea
Stewart R, Rambo TB (2000) Cloacal microbes in house sparrows. Condor 102:679–684. doi:10.1650/0010-5422(2000)102[0679:CMIHS]2.0.CO;2
White J, Mirleau P, Danchin E, Mulard H, Hatch SA, Heeb P, Wagner RH (2010) Sexually transmitted bacteria affect female cloacal assemblages in a wild bird. Ecol Lett 13:1515–1524. doi:10.1111/j.1461-0248.2010.01542.x
van der Hoeven-Hangoor E, van der Vossen JMBM, Schuren FHJ, Verstegen MWA, de Oliveira JE, Montijn RC, Hendriks WH (2013) Ileal microbiota composition of broilers fed various commercial diet compositions. Poult Sci 92:2713–2723. doi:10.3382/ps.2013-03017
Ruiz-de-Castañda R, Vela AI, Lobato E, Briones V, Moreno J (2011) Prevalence of potentially pathogenic culturable bacteria on eggshells and in cloacae of female Pied Flycatchers in a temperate habitat in central Spain. J Field Ornithol 82:215–224
Shawkey MD, Firestone MK, Brodie EL, Beissinger SR (2009) Avian incubation inhibits growth and diversification of bacterial assemblages on eggs. PLoS One 4:e4522. doi:10.1371/journal.pone.0004522
Mead GC (1989) Microbes of the avian cecum: types present and substrates utilized. J Exp Zool Suppl 3:48–54
Kohl K (2012) Diversity and function of the avian gut microbiota. J Comp Physiol B 182:591–602. doi:10.1007/s00360-012-0645-z
Józefiak D, Rutkowski A, Martin SA (2004) Carbohydrate fermentation in the avian ceca: a review. Anim Feed Sci Technol 113:1–15. doi:10.1016/j.anifeedsci.2003.09.007
Waite DW, Taylor MW (2014) Characterizing the avian gut microbiota: membership, driving influences, and potential function. Front Microbiol 5:223. doi:10.3389/fmicb.2014.00223
Yang J (2012) Influence of dietary fibers and whole grains on fecal microbiota during in vitro fermentation. Thesis, the University of Nebraska
Dethlefsen L, Relman DA (2011) Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci 108:4554–4561. doi:10.1073/pnas.1000087107
Ridlon JM, Kang D-J, Hylemon PB (2006) Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. doi:10.1194/jlr.R500013-JLR200
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto J-M, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD, Wang J (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi:10.1038/nature08821
Kita K, Ken IR, Akamine C, Kawada W, Shimura Y, Inamoto T (2014) Influence of propolis residue on the bacterial flora in the cecum of Nanbu Kashiwa. J Poult Sci 51:275–280. doi:10.2141/jpsa.0130137
Dewhirst FE, Paster BJ, Tzellas N, Coleman B, Downes J, Spratt DA, Wade WG (2001) Characterization of novel human oral isolates and cloned 16S rDNA sequences that fall in the family Coriobacteriaceae: description of olsenella gen. nov., reclassification of Lactobacillus uli as Olsenella uli comb. nov. and description of Olsenella profusa sp. nov. Int J Syst Evol Microbiol 51:1797–1804. doi:10.1099/00207713-51-5-1797
Apajalahti JHA, Kettunen A, Bedford MR, Holben WE (2001) Percent G+C profiling accurately reveals diet-related differences in the gastrointestinal microbial community of broiler chickens. Appl Environ Microbiol 67:5656–5667. doi:10.1128/aem.67.12.5656-5667.2001
Oakley BB, Lillehoj HS, Kogut MH, Kim WK, Maurer JJ, Pedroso A, Lee MD, Collett SR, Johnson TJ, Cox NA (2014) The chicken gastrointestinal microbiome. FEMS Microbiol Lett 360:100–112. doi:10.1111/1574-6968.12608
Mizrahi-Man O, Davenport ER, Gilad Y (2013) Taxonomic classification of bacterial 16S rRNA genes using short sequencing reads: evaluation of effective study designs. PLoS One 8:e53608. doi:10.1371/journal.pone.0053608
Acknowledgments
We thank Susan Hammerly for early discussions concerning the feasibility of this study and Shannon Nodolf, DVM, for conducting the sampling. This work was supported by the National Science Foundation (DEB 0948787) to JAJ and U.S. Fish and Wildlife Service to JAJ.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 20 kb)
Rights and permissions
About this article
Cite this article
Zhang, Y., Simon, S.E., Johnson, J.A. et al. Spatial Microbial Composition Along the Gastrointestinal Tract of Captive Attwater’s Prairie Chicken. Microb Ecol 73, 966–977 (2017). https://doi.org/10.1007/s00248-016-0870-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0870-1