Abstract
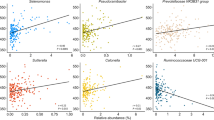
Birds and other animals live and evolve in close contact with millions of microorganisms (microbiota). While the avian microbiota has been well characterized in domestic poultry, the microbiota of other bird species has been less investigated. The aim of this study was to describe the fecal bacterial communities of pet birds. Pooled fecal samples from 22 flocks representing over 150 individual birds of three different species (Melopsittacus undulatus or budgerigars, Nymphicus hollandicus or cockatiels, and Serinus canaria or domestic canaries) were used for analysis using the 16S rRNA gene sequencing in the MiSeq platform (Illumina). Firmicutes was the most abundant phylum (median 88.4 %; range 12.9–98.4 %) followed by other low-abundant phyla such as Proteobacteria (median 2.3 %; 0.1–85.3 %) and Actinobacteria (median 1.7 %; 0–18.3 %). Lactobacillaceae (mostly Lactobacillus spp.) was the most abundant family (median 78.1 %; 1.4–97.5 %), especially in budgerigars and canaries, and it deserves attention because of the ascribed beneficial properties of lactic acid bacteria. Importantly, feces from birds contain intestinal, urinary, and reproductive-associated microbiota thus posing a serious problem to study one anatomical region at a time. Other groups of interest include the family Clostridiaceae that showed very low abundance (overall median <0.1 %) with the exception of two samples from cockatiels (14 and 45.9 %) and one sample from budgerigars (19.9 %). Analysis of UniFrac metrics showed that overall, the microbial communities from the 22 flocks tended to cluster together for each bird species, meaning each species shed distinctive bacterial communities in feces. This descriptive analysis provides insight into the fecal microbiota of pet birds.



Similar content being viewed by others
References
McFall-Ngai M, Hadfield MG, Bosch TCG et al (2013) Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci 110:3229–3236. doi:10.1073/pnas.1218525110
Clemente JC, Ursell LK, Wegener Parfrey L, Knight R (2012) The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. doi:10.1016/j.cell.2012.01.035
Lee WJ, Hase K (2014) Gut microbiota—generated metabolites in animal health and disease. Nat Chem Biol 10:416–424. doi:10.1038/nchembio.1535
Giraud A, Matic I, Tenaillon O et al (2001) Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291:2606–2608. doi:10.1126/science.1056421
Sanchez-Perez G, Mira A, Nyiro G, Pasic L, Rodriguez-Valera F (2008) Adapting to environmental changes using specialized paralogs. Trends Genet 24:154–158. doi:10.1016/j.tig.2008.01.002
Ley RE, Hamady M, Lozupone C et al (2008) Evolution of mammals and their gut microbes. Science 320:1647–1651. doi:10.1126/science.1155725
Slack E, Hapfelmeier S, Stecher B et al (2009) Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 325:617–620. doi:10.1126/science.1172747
Gill F, Donsker D (2016) IOC world bird list (version 6.2). doi: 10.14344/IOC.ML.6.2
Zhang G, Cai L, Li Q et al (2014) Comparative genomics reveal insight into avian genome evolution and adaptation. Science 346:1311–1320. doi:10.1126/science.1251385
Choi KY, Lee TK, Sul WJ (2015) Metagenomic analysis of chicken gut microbiota for improving metabolism and health of chickens—a review. Asian Australas J Anim Sci 28:1217–1225. doi:10.5713/ajas.15.0026
Fujisawa T, Shirasaka S, Watabe J, Mitsuoka T (1984) Lactobacillus aviarius sp. nov.: a new species isolated from the intestine of chickens. Syst Appl Microbiol 5:414–420. doi:10.1016/S0723-2020(84)80042-9
Lan PT, Hayashi H, Sakamoto M, Benno Y (2002) Phylogenetic analysis of cecal microbiota in chicken by the use of 16S rDNA clone libraries. Microbiol Immunol 46:371–382. doi:10.1111/j.1348-0421
Lan PT, Sakamoto M, Benno Y (2004) Effects of two probiotic Lactobacillus strains on jejunal and cecal microbiota of broiler chicken under acute heat stress condition as revealed by molecular analysis of 16S rRNA genes. Microbiol Immunol 48:917–929. doi:10.1111/j.1348-0421
Ludvigsen J, Svihus B, Rudi K (2016) Rearing room affects the non-dominant chicken cecum microbiota, while diet affects the dominant microbiota. Front Vet Sci 3:16. doi:10.3389/fvets
Meng H, Zhang Y, Zhao L et al (2014) Body weight selection affects quantitative genetic correlated responses in gut microbiota. PLoS ONE 9:e89862. doi:10.1371/journal.pone
Neumann AP, Suen G (2015) Differences in major bacterial populations in the intestine of mature broilers after feeding virginiamycin or bacitracin methylene disalicylate. J Appl Microbiol 119:1515–1526. doi:10.1111/jam.12960
Oakley BB, Lillehoj HS, Kogut MH et al (2014) The chicken gastrointestinal microbiome. FEMS Microbiol Lett 360:100–112. doi:10.1111/1574-6968
Pan D, Yu Z (2014) Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes 5:108–119. doi:10.4161/gmic.26945
Polansky O, Sekelova Z, Faldynova M, Sebkova A, Sisak F, Rychlik I (2015) Important metabolic pathways and biological processes expressed by chicken cecal microbiota. Appl Environ Microbiol 82:1569–1576. doi:10.1128/AEM.03473-15
Stanley D, Hughes RJ, Moore RJ (2014) Microbiota of the chicken gastrointestinal tract: influence on health, productivity and disease. Appl Microbiol Biotechnol 98:4301–4310. doi:10.1007/s00253-014-5646-2
Witzig M, Camarinha da Silva A, Green-Engert R et al (2015) Spatial variation of the gut microbiota in broiler chickens as affected by dietary available phosphorus and assessed by T-RFLP analysis and 454 pyrosequencing. PLoS ONE 10:e0143442. doi:10.1371/journal.pone
Yeoman CJ, Chia N, Jeraldo P, Sipos M, Goldenfeld ND, White BA (2012) The microbiome of the chicken gastrointestinal tract. Anim Health Res Rev 13:89–99. doi:10.1017/S1466252312000138
Zhao L, Wang G, Siegel P et al (2013) Quantitative genetic background of the host influences gut microbiomes in chickens. Sci Rep 3:1163. doi:10.1038/srep01163
Benskin CMH, Rhodes G, Pickup RW et al (2015) Life history correlates of fecal bacterial species richness in a wild population of the blue tit Cyanistes caeruleus. Ecol Evol 5:821–835. doi:10.1002/ece3.1384
Jacob S, Parthuisot N, Vallat A, Ramon-Portugal F, Helfenstein F, Heeb P (2015) Microbiome affects egg carotenoid investment, nestling development and adult oxidative costs of reproduction in Great tits. Funct Ecol 29:1048–1058. doi:10.1111/1365-2435.12404
Lawson PA, Wacher C, Hansson I, Falsen E, Collins MD (2001) Lactobacillus psittaci sp. nov., isolated from a hyacinth macaw. Int J Syst Evol Microbiol 51:967–970. doi:10.1099/00207713-51-3-967
Splichalova P, Svec P, Ghosh A et al (2015) Prevalence, diversity and characterization of enterococci from three coraciiform birds. Antonie Van Leeuwenhoek 107:1281–1289. doi:10.1007/s10482-015-0422-6
van Dongen WFD, White J, Brandl HB et al (2013) Age-related difference in the cloacal microbiota of a wild bird species. BMC Ecol 13:11. doi:10.1186/1472-6785-13-11
Xenoulis PG, Gray PL, Brightsmith DJ et al (2010) Molecular characterization of the cloacal microbiota of healthy wild and captive parrots. Vet Microbiol 15:320–325. doi:10.1016/j.vetmic.2010.05.024
Berkhoff H (1985) Clostridium colinum sp. nov., nom. rev., the causative agent of ulcerative enteritis (quail disease) in quail, chickens, and pheasants. Int J Syst Bacteriol 35:155–159. doi:10.1099/00207713-35-2-155
Brilhante RSN, Castelo-Branco DSCM, Soares GDP et al (2010) Characterization of the gastrointestinal yeast microbiota of cockatiels (Nymphicus hollandicus): a potential hazard to human health. J Med Microbiol 59:718–723. doi:10.1099/jmm.0.017426-0
Cooper KK, Songer JG, Uzal FA (2013) Diagnosing clostridial enteric disease in poultry. J Vet Diagn Investig 25:314–327. doi:10.1177/1040638713483468
Griekspoor P, Colles FM, McCarthy ND et al (2013) Marked host specificity and lack of phylogeographic population structure of Campylobacter jejuni in wild birds. Mol Ecol 22:1463–1472. doi:10.1111/mec.12144
Perelman B, Mints S, Zjut M, Kuttin E, Machny S (1991) An unusual Clostridium colinum infection in broiler chickens. Avian Pathol 20:475–480. doi:10.1080/03079459108418785
Suchodolski JS, Markel ME, Garcia-Mazcorro JF, Unterer S, Heilmann RM, Dowd SE (2012) The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS ONE 7:e51907. doi:10.1371/journal.pone
Caporaso JG, Lauber CL, Walters WA et al (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi:10.1038/ismej.2012.8
Garcia-Mazcorro JF, Ivanov I, Mills DA, Noratto G (2016) Influence of whole-wheat consumption on fecal microbial ecology of obese diabetic mice. PeerJ 4:e1702. doi:10.7717/peerj.1702
Garcia-Mazcorro JF, Mills DA, Noratto G (2016) Molecular exploration of fecal microbiome in quinoa supplemented obese mice. FEMS Microbiol Ecol. doi:10.1093/femsec/fiw089
Caporaso JG, Kuczynski J, Stombaugh J et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi:10.1038/nmeth.f.303
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi:10.1093/bioinformatics/btq461
Rideout JR, He Y, Navas-Molina JA et al (2014) Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2:e545. doi:10.7717/peerj.545
Langille MG, Zaneveld J, Caporaso JG et al (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–823. doi:10.1038/nbt.2676
Tikhonov M, Leach RW, Wingreen NS (2015) Interpreting 16S metagenomic data without clustering to achieve sub-OTU resolution. ISME J 9:68–80. doi:10.1038/ismej.2014.117
Nguyen NP, Warnow T, Pop M, White B (2016) A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity. npj Biofilms Microbiomes 2:16004. doi:10.1038/npjbiofilms.2016.4
Chen W, Zhang CK, Cheng Y, Zhang S, Zhao H (2013) A comparison of methods for clustering 16S rRNA sequences into OTUs. PLoS ONE 8:e70837. doi:10.1371/journal.pone.0070837
He Y, Caporaso JG, Jiang XT et al (2015) Stability of operational taxonomic units: an important but neglected property for analyzing microbial diversity. Microbiome 3:20. doi:10.1186/s40168-015-0081-x
Westcott SL, Schloss PD (2015) De novo clustering methods outperform reference-based methods for assigning 16S rRNA gene sequences to operational taxonomic units. PeerJ 3:e1487. doi:10.7717/peerj.1487
Koeppel AF, Wu M (2013) Surprisingly extensive mixed phylogenetic and ecological signals among bacterial operational taxonomic units. Nucleic Acids Res 41:5175–5188. doi:10.1093/nar/gkt241
Garcia-Mazcorro JF (2013) Testing evolutionary models to explain the process of nucleotide substitution in gut bacterial 16S rRNA gene sequences. FEMS Microbiol Lett 346:97–104. doi:10.1111/1574-6968.12207
Garcia-Mazcorro JF, Barcenas-Walls JR (2016) Thinking beside the box: should we care about the noncoding strand of the 16S rRNA gene? FEMS Microbiol Lett. doi:10.1093/femsle/fnw171
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27:379–423
Faith DP, Baker AM (2006) Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol Bioinforma 2:121–128
Chao A (1984) Nonparametric-estimation of the number of classes in a population. Scand J Stat 11:265–270
Bent SJ, Forney LJ (2008) The tragedy of the uncommon: understanding limitations in the analysis of microbial diversity. ISME J 2:689–695. doi:10.1038/ismej.2008.44
Lozupone CA, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. doi:10.1128/AEM.01996-06
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Paleontol Electron 4:1–9
Parks DH, Beiko RG (2010) Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26:715–721. doi:10.1093/bioinformatics/btq041
R Development Core Team (2008) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org
Lukjancenko O, Wassenaar TM, Ussery DW (2010) Comparison of 61 sequenced Escherichia coli genomes. Microb Ecol 60:708–720. doi:10.1007/s00248-010-9717-3
Jaspers E, Overmann J (2004) Ecological significance of microdiversity: identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl Environ Microbiol 70:4831–4839. doi:10.1128/AEM.70.8.4831-4839
Plassart C, Mauvais F, Heurté J, Sautereau J, Legeay C, Bouvet P (2013) First case of intra-abdominal infection with Clostridium disporicum. Anaerobe 19:77–78. doi:10.1016/j.anaerobe.2012.12.002
Gao J, Liu G, Li H, Xu L, Du L, Yang B (2016) Predictive functional profiling using marker gene sequences and community diversity analyses of microbes in full-scale anaerobic sludge digesters. Bioprocess Biosyst Eng
Tyx RE, Stanfill SB, Keong LM, Rivera AJ, Satten GA, Watson CH (2016) Characterization of bacterial communities in selected smokeless tobacco products using 16S rDNA analysis. PLoS ONE 11:e0146939. doi:10.1371/journal.pone
Cisek AA, Binek M (2014) Chicken intestinal microbiota function with a special emphasis on the role of probiotic bacteria. Pol J Vet Sci 17:385–394. doi:10.2478/pjvs-2014-0057
Waite DW, Taylor MW (2014) Characterizing the avian gut microbiota: membership, driving influences, and potential function. Front Microbiol 5:1–12. doi:10.3389/fmicb.2014.00223
Wu GD, Chen J, Hoffmann C et al (2011) Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi:10.1126/science.1208344
Hoyles L, McCartney AL (2009) What do we mean when we refer to Bacteroidetes populations in the human gastrointestinal tract? FEMS Microbiol Lett 299:175–183. doi:10.1111/j.1574-6968.2009.01741
Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW (2011) Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35:652–680. doi:10.1111/j.1574-6976.2011.00269.x
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi:10.1128/AEM.71.12.8228-8235.2005
Campbell AM, Fleisher J, Sinigalliano C, White JR, Lopez JV (2015) Dynamics of marine bacteria community diversity of the coastal waters of the reefs, inlets, and wastewater outfalls of southeast Florida. Microbiol Open 4:390–408. doi:10.1002/mbo3.245
Igarashi H, Maeda S, Ohno K, Horigome A, Odamaki T, Tsujimoto H (2014) Effect of oral administration of metronidazole or prednisolone on fecal microbiota in dogs. PLoS ONE 9:e107909. doi:10.1371/journal.pone.0107909
Wu GD, Lewis JD, Hoffmann C et al (2010) Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol 10:206. doi:10.1186/1471-2180-10-206
Lee WY (2015) Avian gut microbiota and behavioral studies. Kor J Orni 22:1–11
Archie EA, Theis KR (2011) Animal behavior meets microbial ecology. Anim Behav 82:425–436
Fischer I, Christen C, Lutz H et al (2006) Effects of two diets on the haematology, plasma chemistry and intestinal flora of budgerigars (Melopsittacus undulatus). Vet Rec 159:480–484
Kohl KD (2012) Diversity and function of the avian gut microbiota. J Comp Physiol B 182:591–602. doi:10.1007/s00360-012-0645-z
Waite DW, Taylor MW (2015) Exploring the avian gut microbiota: current trends and future directions. Front Microbiol 6:673. doi:10.3389/fmicb.2015.00673
Fricke WF, Maddox C, Song Y, Bromberg JS (2014) Human microbiota characterization in the course of renal transplantation. Am J Transplant 14:416–427. doi:10.1111/ajt.12588
Acknowledgments
Part of this study was presented at ISME15 in Seoul, South Korea. JFGM acknowledges financial support by PRODEP (Secretaria de Educacion Publica, Mexico, grant number DSA/103.5/14/11021). The authors also wish to thank the QIIME and PICRUSt developers and users for their valuable help through the QIIME and PICRUSt Forums.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 387 kb)
Rights and permissions
About this article
Cite this article
Garcia-Mazcorro, J.F., Castillo-Carranza, S.A., Guard, B. et al. Comprehensive Molecular Characterization of Bacterial Communities in Feces of Pet Birds Using 16S Marker Sequencing. Microb Ecol 73, 224–235 (2017). https://doi.org/10.1007/s00248-016-0840-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0840-7