Abstract
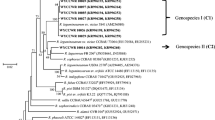
Aiming to investigate the diversity and distribution of rhizobia associated with Ammopiptanthus, an endangered evergreen legume widely distributed in deserts, we characterized a total of 219 nodule isolates from nine sampling sites in Northwest China with different soil characteristics based upon restriction fragment length polymorphism (RFLP) analysis of 16S ribosomal RNA (rRNA) and symbiotic genes (nodC and nifH). Ten isolates representing different 16S rRNA-RFLP types were selected for further sequence analyses of 16S rRNA and four housekeeping genes. As results, nine genospecies belonging to the genera Ensifer, Neorhizobium, Agrobacterium, Pararhizobium, and Rhizobium could be defined among the isolates. The nodC and nifH phylogenies of 14 isolates representing different symbiotic-RFLP types revealed five lineages linked to Ensifer fredii, Ensifer meliloti, Rhizobium leguminosarum, Mesorhizobium amorphae, and Rhizobium gallicum, which demonstrated the various origins and lateral transfers of symbiotic genes between different genera and species. The rhizobial diversities of Ammopiptanthus mongolicus varied among regions, and the community compositions of rhizobia associated with A. mongolicus were significantly different in wild and cultured fields. Constrained correspondence analysis showed that the distribution of A. mongolicus rhizobia could be explained by available potassium content and that the assembly of symbiotic types was mainly affected by available phosphorus content and carbon–nitrogen ratio.



Similar content being viewed by others
References
Liu JQ, Qiu XM, Yang K, Shi QH (1995) Studies on the plant community of Ammopiptanthus mongolicus. J Desert Res 15:109–115 (in Chinese with English abstract)
Ge XJ, Yu Y, Yuan YM, Huang HW, Yan C (2005) Genetic diversity and geographic differentiation in endangered Ammopiptanthus (Leguminosae) populations in desert regions of northwest China as revealed by ISSR analysis. Ann Bot 95:843–851
Liu GH (1998) Study on the endangered reasons of Ammopiptanthus mongolicus in the desert of Alashan. Bull Bot Res 18:341–345
Cheng SH (1959) Ammopiptanthus Cheng f., a new genus of Leguminosae from Central Asia. J Bot USSR 44:1381–1386
Xu S, An L, Feng H, Wang X, Li X (2002) The seasonal effects of water stress on Ammopiptanthus mongolicus in a desert environment. J Arid Environ 51:437–447
Pang T, Ye CY, Xia XL, Yin WL (2013) De novo sequencing and transcriptome analysis of the desert shrub, Ammopiptanthus mongolicus, during cold acclimation using Illumina/Solexa. BMC Genomics 14:488
Sprent JI (2009) Legume nodulation, a global respective. p 117. Willey-Blackwell, Singapore. http://books.google.com.hk/books?isbn=1444316397
He H, Wang H, Jia G (2012) Nodule histology and ultrastructure of Ammopiptanthus mongolicus and subcellular localization of glycoprotein in nodules. Sci Silvae Sin 48:31–38 (in Chinese with English abstract)
Bi JT, He DH, Chen WM, Xie RM, Wei GH (2009) Analysis of genetic diversity and phylogenesis of rhizobia from Ammopiptanthus mongolicus. Acta Botan Boreali-Occiden Sin 29:695–703 (in Chinese with English abstract)
Bi J, He D, Xie R, Wei G (2009) PCR-RFLP analysis of nodulation gene nodA of rhizobia from Ammopiptanthus mongolicus. J Desert Res 29:703–710 (in Chinese with English abstract)
Bi J, He D, Xie R, Wei X, Wei G (2009) PCR-RFLP analysis of nitrogenase gene nifH of rhizobia from Ammopiptanthus mongolicus. Acta Agric Bor-Accid Sin 18:315–321 (in Chinese with English abstract)
Han LL, Wang ET, Han TX, Liu J, Sui XH, Chen WF, Chen WX (2009) Unique community structure and biogeography of soybean rhizobia in the saline–alkaline soils of Xinjiang, China. Plant Soil 324:291–305
Li L, Hanna S, Leone M, Wei GH, Lindström K, Räsänen LA (2011) Biogeography of symbiotic and other endophytic bacteria isolated from medicinal Glycyrrhiza species in China. FEMS Microbial Ecol 79:46–68
Li QQ, Wang ET, Zhang YZ, Zhang YM, Tian CF, Sui XH et al (2011) Diversity and biogeography of rhizobia isolated from root nodules of Glycine max grown in Hebei province of China. Microbial Ecol 61:917–931
Zhao L, Fan MC, Zhang DH, Yang RP, Zhang FL, Xu L et al (2014) Distribution and diversity of rhizobia associated with wild soybean (Glycine soja Sieb. & Zucc.) in Northwest China. Syst Appl Microbiol 37:449–456
Zhao LF, Deng ZS, Yang WQ, Cao Y, Wang ET, Wei GH (2010) Diverse rhizobia associated with Sophora alopecuroides grown in different regions of Loess Plateau in China. Syst Appl Microbiol 33:468–477
Zhang YM, Li Y, Chen WF, Wang ET, Tian CF, Li QQ, Zhang YZ, Sui XH, Chen WX (2011) Biodiversity and biogeography of rhizobia associated with soybean plants grown in the North China Plain. Appl Environ Microbiol 77:6331–6342
Vincent JM (1970) A manual for the practical study of root-nodule bacteria. IBP handbook no. 15. International Biological Programme, London
Afnor (2005) Soil quality, determination of pH. NF ISO 10390. AFNOR, Paris, France
Houba VJG, Novozamsky I, Huybregts AWM, Van der Lee J (1986) Comparison of soil extractions by 0.01M CaCl2, by EUF and by some conventional extraction procedures. Plant Soil 96:433–437
Olsen SR (1954) Estimation of available phosphorus in soils by extraction with sodium bicarbonate. USDA, Washington, DC
Simonis AD (1996) Effect of temperature on extraction of phosphorus and potassium from soils by various extracting solutions. Commun Soil Sci Plant Anal 27:665–684
Cao Y, Wang ET, Zhao L, Chen WM, Wei GH (2014) Diversity and distribution of rhizobia nodulated with Phaseolus vulgaris in two regions of China. Soil Biol Biochem 78:128–137
Tan ZY, Xu XD, Wang ET, Gao JL, Martinez-Romero E, Chen WX (1997) Phylogenetic and genetic relationships of Mesorhizobium tianshanense and related rhizobia. Int J Syst Evol Microbiol 47:874–879
Pinto FGS, Hungria M, Mercante FM (2007) Polyphasic characterization of Brazilian Rhizobium tropici strains effective in fixing N2 with common bean (Phaseolus vulgaris L.). Soil Biol Biochem 39:1851–1864
Yang WQ, Kong ZY, Chen WM, Wei GH (2013) Genetic diversity and symbiotic evolution of rhizobia from root nodules of Coronilla varia. Syst Appl Microbiol 36:49–55
Martens M, Dawyndt P, Coopman R, Gillis M, De Vos P, Willems A (2007) Multilocus sequence analysis of Ensifer and related taxa. Int J Syst Evol Microbiol 57:489–503
Martens M, Dawyndt P, Coopman R, Gillis M, De Vos P, Willems A (2008) Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int J Syst Evol Microbiol 58:200–214
Mousavi SA, Osterman J, Wahlberg N, Nesme X, Lavire C, Vial L et al (2014) Phylogeny of the Rhizobium–Allorhizobium–Agrobacterium clade supports the delineation of Neorhizobium gen. nov. Syst Appl Microbiol 37:208–215
Vinuesa P, Silva C, Lorite MJ, Izaguirre-Mayoral ML, Bedmar EJ, Martinez-Romero E (2005) Molecular systematics of rhizobia based on maximum likelihood and Bayesian phylogenies inferred from rrs, atpD, recA and nifH sequences, and their use in the classification of Sesbania microsymbionts from Venezuelan wetlands. Syst Appl Microbiol 28:702–716
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Grant T, Kluge AG (2003) Data exploration in phylogenetic inference: scientific, heuristic, or neither. Cladistics Int J Willi Hennig Soc 19:379–418
Xia XH (2009) Assessing substitution saturation with DAMBE. In: Lemey P, Salemi M, Vandamme AM (eds) The phylogenetic handbook. Cambridge University Press, Cambridge, pp 615–630
Xia XH (2013) DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol 30:1720–1728
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Swofford D (2002) PAUP*: phylogenetic analysis using parsimony and other methods. Version 4. Sinauer Associates, Sunderland, MA
Huelsenbeck JP, Ronquist FR (2011) MrBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Ronquist FR, Huelsenbeck JP (2003) MrBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Guindon S, Gascuel O (2003) PhyML: a simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Farris JS, Källersjö M, Kluge AG, Bult C (1994) Testing significance of incongruence. Cladistics Int J Willi Hennig Soc 10:315–319
Farris JS, Källersjö M, Kluge AG, Bult C (1995) Constructing a significance test for incongruence. Syst Biol 44:570–572
Legendre P, Legendre L (1998) Numerical ecology, 2nd English edn. Elsevier Science BV, Amsterdam
ter Braak CJF (1986) Canonical correspondence analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67:1167–1179
Mousavi SA, Willems A, Nesme X, de Lajudie P, Lindström K (2015) Revised phylogeny of Rhizobiaceae: proposal of the delineation of Pararhizobium gen. nov., and 13 new species combinations. Syst Appl Microbiol 38:84–90
Berkum PV, Terefework Z, Paulin L, Suomalainen S, Lindström K, Eardly BD (2003) Discordant phylogenies within the rrn loci of rhizobia. J Bacteriol 185:2988–2998
Eardly BD, Nour SA, van Berkum P, Selander RK (2005) Rhizobial 16S rRNA and dnaK genes: mosaicism and the uncertain phylogenetic placement of Rhizobium galegae. Appl Environ Microbiol 71:1328–1335
Silva C, Vinuesa P, Eguiarte LE, Souza V, Martinez-Romero E (2005) Evolutionary genetics and biogeographic structure of Rhizobium gallicum sensu lato, a widely distributed bacterial symbiont of diverse legumes. Mol Ecol 14:4033–4050
Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N (2001) Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology UK 147:981–993
Sullivan JT, Ronson CW (1998) Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci U S A 95:5145–5149
Sullivan JT, Patrick HN, Lowther WL, Scott DB, Ronson CW (1995) Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc Natl Acad Sci U S A 92:8985–8989
Sullivan JT, Trzebiatowski JR, Cruickshank RW, Gouzy J, Brown SD, Elliot RM et al (2002) Comparative sequence analysis of the symbiosis island of Mesorhizobium lotis strain R7A. J Bacteriol 184:3086–3095
Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S et al (2002) Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res 9:189–197
Lindström K (1989) Rhizobium galegae, a new species of legume root nodule bacteria. Int J Syst Bacteriol 39:365–367
Radeva G, Jürgens G, Niemi M, Nick G, Suominen L, Lindström K (2001) Description of two biovars in the Rhizobium galegae species: biovar orientalis and biovar officinalis. Syst Appl Microbiol 24:192–205
Bailly X, Olivieri I, Brunel B, Cleyet-Marel JC, Béna G (2007) Horizontal gene transfer and homologous recombination drive the evolution of the nitrogen-fixing symbionts of Medicago species. J Bacteriol 189:5223–5236
Marchetti M, Capela D, Glew M, Cruveiller S, Chane-Woon-Ming B, Gris C et al (2010) Experimental evolution of a plant pathogen into a legume symbiont. PLoS Biol 8:1–10
Fernández-Calviño D, Baath E (2010) Growth response of the bacterial community to pH in soils differing in pH. FEMS Microbiol Ecol 73:149–156
Rousk J, Baath E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG et al (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351
Appunu C, N’Zoue A, Laguerre G (2008) Genetic diversity of native bradyrhizobia isolated from soybeans (Glycine max L.) in different agricultural–ecological–climatic regions of India. Appl Environ Microbiol 74:5991–5996
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103:626–631
Bonilla I, Bolãnos L (2009) Mineral nutrition for legume–rhizobia symbiosis: B, Ca, N, P, S, K, Fe, Mo, Co, and Ni: a review. Sustainable agriculture reviews. Org Farm Pest Control Remediat Soil Pollut 1:253–274
Acknowledgments
We thank Zhengshan Liu and Peng Zhao for helping in nodule collection. This work was supported by project from the National Science Foundation of China (31125007).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1570 kb)
Rights and permissions
About this article
Cite this article
Zhao, L., Wang, X., Huo, H. et al. Phylogenetic Diversity of Ammopiptanthus Rhizobia and Distribution of Rhizobia Associated with Ammopiptanthus mongolicus in Diverse Regions of Northwest China. Microb Ecol 72, 231–239 (2016). https://doi.org/10.1007/s00248-016-0759-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0759-z