Abstract
Isabel Lake is a moderate saline soda crater lake located in Isabel Island in the eastern tropical Pacific coast of Mexico. Lake is mainly formed by rainfall and is strongly affected by evaporation and high input of nutrients derived from excretions of a large bird community inhabiting the island. So far, only the island macrobiota has been studied. The knowledge of the prokaryotic biota inhabiting the upper layers of this meromictic lake can give clues for the maintenance of this ecosystem. We assessed the diversity and composition of prokaryotic community in sediments and water of the lake by DGGE profiling, 16S rRNA gene amplicon pyrosequencing, and cultivation techniques. The bacterial community is largely dominated by halophilic and halotolerant microorganisms. Alpha diversity estimations reveal higher value in sediments than in water (P > 0.005). The lake water is dominated by γ-Proteobacteria belonging to four main families where Halomonadaceae presents the highest abundance. Aerobic, phototrophic, and halotolerant prokaryotes such as Cyanobacteria GPIIa, Halomonas, Alcanivorax, Idiomarina, and Cyclobacterium genera are commonly found. However, in sediment samples, Formosa, Muricauda, and Salegentibacter genera corresponding to Flavobacteriaceae family accounted for 15–20 % of the diversity. Heterotrophs like those involved in sulfur cycle, Desulfotignum, Desulfuromonas, Desulfofustis, and Desulfopila, appear to play an important role in sediments. Finally, a collection of aerobic halophilic bacterial isolates was created from these samples; members of the genus Halomonas were predominantly isolated from lake water. This study contributes to state the bacterial diversity present in this particular soda saline crater lake.



Similar content being viewed by others
References
Ortega GF, González GR (1980) Nódulos de peridotita en la isla Isabel, Nayarit. Universidad Nacional Autónoma de México. Rev Mex Cienc Geol 4:82–83
Sorokin DY, Berben T, Melton ED, Overmars L, Vavourakis CD, Muyzer G (2014) Microbial diversity and biogeochemical cycling in soda lakes. Extremophiles 18:791–809
Hahn IJ, Hogeback S, Romer U, Vergara PM (2012) Biodiversity and biogeography of birds in Pacific Mexico along an isolation gradient from mainland Chamela via coastal Marias to oceanic Revillagigedo Islands. Vertebr Zool 62:123–144
Alcocer J, Lugo A, Sánchez MR, Escobar E (1998) Isabela Crater-Lake: a Mexican insular saline lake. Hydrobiologia 381:1–7
Diario Oficial de la Federación (1980). Decreto por el que se declara Parque Nacional a la Isla Isabel, ubicada frente a las costas del Estado de Nayarit, declarándose de interés público la conservación y aprovechamiento de sus valores naturales, para fines recreativos, culturales y de investigación científica. http://dof.gob.mx/nota_detalle.php?codigo=4862220&fecha=08/12/1980
Grant WD (1992) Alkaline environments. In: Lederberg journal of encyclopedia of microbiology. Academic Press, London 1: 73–80
Mesbah NM, Abou-El-Ela SH, Wiegel J (2007) Novel and unexpected prokaryotic diversity in water and sediments of the alkaline, hypersaline lakes of the Wadi An Natrun, Egypt. Microb Ecol 54:598–617
Hollister EB, Engledow AS, Hammett AJ, Provin TL, Wilkinson HH, Gentry TJ (2010) Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME J 4:829–838
Tourova TP, Slobodova NV, Bumazhkin BK, Kolganova TV, Muyzer G, Dimitry Sorokin DY (2012) Analysis of community composition of sulfur-oxidizing bacteria in hypersaline and soda lakes using soxB as a functional molecular marker. FEMS Microbiol Ecol 84:280–289
Zavarzin GA, Zhilina TN, Kevbrin VV (1999) The alkaliphilic microbial community and its functional diversity. Microbiology 68:503–521
Rodríguez-Valera F (1988) Characteristics and microbial ecology of hypersaline environments. In: Rodríguez-Valera F (ed) Halophilic bacteria, Ith edn. CRC Press, Boca Raton, FL, pp 3–30
Ventosa A, Nieto JJ, Oren A (1998) Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev 62:504–544
Jiang H, Dong H, Zhang G, Yu B, Chapman LR, Fields MW (2006) Microbial diversity in water and sediment of Lake Chaka, an athalasso haline lake in Northwestern China. Appl Environ Microbiol 72:3832–3845
Ghai R, Pašić L, Fernández AB, Martin-Cuadrado AB, Mizuno CM, McMahon KD, Papke RT, Stepanauskas R, Rodriguez-Brito B, Rohwer F, Sánchez-Porro C, Ventosa A, Rodríguez-Valera F (2011) New abundant microbial groups in aquatic hypersaline environments. Sci Rep 1:135
Lozupone CA, Knight R (2007) Global patterns in bacterial diversity. Proc Natl Acad Sci U S A 104:11436–11440
Canfora L, Bacci G, Pinzari F, Lo Papa G, Dazzi C, Benedetti A (2014) Salinity and bacterial diversity: to what extent does the concentration of salt affect the bacterial community in a saline soil? PLoS One 9:e106662
Dimitriu PA, Pinkart HC, Peyton BM, Mormile MR (2008) Spatial and temporal patterns in the microbial diversity of a meromictic soda lake in Washington State. Appl Environ Microbiol 74:4877–4888
Pagaling E, Wang H, Venables M, Wallace A, Grant WD, Cowan DA, Jones BE, Ma Y, Ventosa A, Heaphy S (2009) Microbial biogeography of six salt lakes in Inner Mongolia, China, and a Salt Lake in Argentina. Appl Environ Microbiol 75:5750–5760
Lanzen A, Simachew A, Gessesse A, Chmolowska D, Jonassen I, Ovreas L (2013) Surprising prokaryotic and eukaryotic diversity, community structure and biogeography of Ethiopian soda lakes. PLoS ONE 8:e72577
Dadheech PK, Glöckner G, Casper P, Kotut K, Mazzoni CJ, Mbedi S, Krienitz L (2013) Cyanobacterial diversity in the hot spring, pelagic and benthic habitats of a tropical soda lake. FEMS Microbiol Ecol 85:389–401
Asao M, Pinkart HC, Madigan MT (2011) Diversity of extremophilic purple phototrophic bacteria in Soap Lake, a central Washington (USA) soda lake. Environ Microbiol 13:2146–2157
de la Haba RR, Arahal DR, Márquez MC, Ventosa A (2010) Phylogenetic relationships within the family Halomonadaceae based on comparative 23S and 16S rRNA gene sequence analysis. Int J Syst Evol Microbiol 60:737–748
de la Haba RR, Sanchez-Porro C, Ventosa A (2011) Taxonomy, phylogeny, and biotechnological interest of the family halomonadaceae. In: Ventosa A, Oren A, Ma Y (eds) Halophiles and hypersaline environments. Springer, Heidelbeg, pp 27–64
Housh TB, Aranda-Gomez JJ, Luhr JF (2010) Isla Isabel (Nayarit, México): quaternary alkali basalts with mantle xenoliths erupted in the mouth of the Gulf of California. J Volcanol Geotherm Res 197:85–107
Romero-Viana L, Kienel U, Sasche D (2012) Lipid biomarker signatures in a hypersaline lake on Isabel island (Eastern Pacific) as a proxy for past rainfall anomaly (1942–2006 AD). Palaeogeogr Palaeoclimatol Palaeoecol 350:49–61
Kempers AJ (1974) Determination of sub-microquantities of ammonium and nitrates in soils with phenol, sodium nitroprusside and hypochlorite. Geoderma 12:201–206
Ramírez-Saad H, Akkermans W, Akkermans ADL (1996) DNA extraction from actinorhizal nodules. Chap. 1.4.4. In: Akkermans ADL, van Elsas JD, de Bruijn F (eds) Manual molecular microbial ecology. Kluwer Academic Publishers, Dordrecht, pp 1–11
Nübel U, Engelen B, Felske A, Snaidr A, Wieshuber A, Amann RI, Ludwig W, Backhaus H (1996) Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol 178:5636–5643
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Ramírez-Saad H, Janse J, Akkermans ADL (1998) Root nodules of Ceanothus caeruleus contain both; the N2-fixing Frankia endophyte and a phylogenetically related Nod−/Fix− actinomycete. Can J Microbiol 44:140–148
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C (1998) Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans ADL, van Elsas JD, de Bruijn FJ (eds) Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, pp 1–27
Sanguinetti CJ, Dias Nieto E, Simpson AJG (1994) Rapid silver staining and recovery of PCR products separated on polyacrilamyde gels. Biotechniques 17:915–919
Ramírez-Saad HC, Sessitsch A, de Vos WM, Akkermans ADL (2000) Bacterial community changes and enrichment of uncultured Burkholderia-like bacteria induced by chlorinated benzoates in a peat-forest soil-microcosm. Syst Appl Microbiol 23:591–598
Clarke KR (1993) Non-parametric multivariate analysis of changes in community structure. Aust J Ecol 18:117–143
Binladen J, Gilbert MT, Bollback JP, Panitz F, Bendixen C, Nielsen R, Willerslev E (2007) The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE 2(2):e197
Baker G, Smith JJ, Cowan DA (2003) Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55:541–555
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. doi:10.1093/bioinformatics/btr381
Flynn JM, Brown EA, Chain FJJ, MacIsaac HJ, Cristescu ME (2015) Toward accurate molecular identification of species in complex environmental samples: testing the performance of sequence filtering and clustering methods. Ecol Evol. doi:10.1002/ece3.1497
Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO (2014) The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–648
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Parks DH, Tyson GW, Hugenholtz P, Beiko RG (2014) STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30:3123–3124
Nguyen NH, Smith D, Peay K, Kennedy P (2015) Parsing ecological signal from noise in next generation amplicon sequencing. New Phytol 205:1389–1393
Antony CP, Kumaresan D, Hunger S, Drake HL, Murrel JC, Shouche YS (2013) Microbiology of Lonar Lake and other soda lakes. ISME J 7:468–476
Culkin F, Smith N (1980) Determination of the concentration of potassium chloride solution having the same electrical conductivity, at 15°C and infinite frequency, as standard seawater of salinity 35.0000 ‰ (Chlorinity 19.37394 ‰). IEEE J Ocean Eng 5:22–23
Grant WD (2006) Alkaline environments and biodiversity, in extremophilies. In: Gerday C, Glansdorff N (eds) Encyclopedia of Life Support Systems (EOLSS). Developed under the Auspices of The UNESCO. Eolss Publishers, Oxford, UK
Shapovalova AA, Hizhniak TV, Tourova TP, Muyzer G, Sorokin DY (2009) Halomonas chromatireducens sp. nov., a novel haloalkaliphile from soda soil capable of aerobic chromate reduction. Microbiology (Moscow) 78:117–127
Bagheri M, Amoozegar MA, Didari M, Makhdoumi-Kakhki A, Schumann P, Spröer C, Sánchez-Porro C, Ventosa A (2013) Marinobacter persicus sp. nov., a moderately halophilic bacterium from a saline lake in Iran. Antonie Van Leeuwenhoek 104:47–54
Nedashkovskaya OI, Kim SB, Vancanneyt M, Shin DS, Lysenko AM, Shevchenko LS, Krasokhin VB, Mikhailov VV, Swings J, Bae KS (2006) Salegentibacter agarivorans sp. nov., a novel marine bacterium of the family flavobacteriaceae isolated from the sponge artemisina sp. Int J Syst Evol Microbiol 56:883–887
Nedashkovskaya OI, Kim SB, Vancanneyt M, Snauwaert C, Lysenko AM, Rohde M, Frolova GM, Zhukova NV, Mikhailov VV, Bae KS, Oh HW, Swings J (2006) Formosa agariphila sp. nov., a budding bacterium of the family Flavobacteriaceae isolated from marine environments, and emended description of the genus Formosa. Int J Syst Evol Microbiol 56:161–167
Bruns A, Rohde M, Berthe-Corti L (2001) Muricauda ruestringensis gen. nov., sp. nov., a facultatively anaerobic, appendaged bacterium from German North Sea intertidal sediment. Int J Syst Evol Microbiol 51:1997–2006
Wani AA, Surakasi VP, Siddharth J, Raghavan RG, Patole MS, Ranade D, Shouche YS (2006) Molecular analyses of microbial diversity associated with the Lonar soda lake in India: an impact crater in a basalt area. Res Microbiol 157:928–937
Wains M, Tindall BJ, Schumann P, lngvorsen K (1999) Gracilibacillus gen. nov., with description of gracilibacillus halotolerans gen. nov., sp. nov.; transfer of bacillus dipsosauri to gracilibacillus dipsosauri comb. nov., and bacillus salexigens to the genus salibacillus gen. nov., as salibacillus salexigens comb. nov. Int J Syst Evol Microbiol 49:821–831
Rees HC, Grant WD, Jones BE, Heaphy S (2003) Diversity of Kenyan soda Lake alkaliphiles assessed by molecular methods. Extremophiles 8:63–71
Desnues C, Michotey VD, Wieland A, Zhizang C, Fourcans A, Duran R, Bonin PC (2007) Seasonal and dial distributions of denitrifying and bacterial communities in a hypersaline microbial mat (Camargue, France). Water Res 41:3407–3419
Foti MJ, Sorokin DY, Zacharova EE, Pimenov NV, Gijs Kuenen JG, Muyzer G (2008) Bacterial diversity and activity along a salinity gradient in soda lakes of the Kulunda Steppe (Altai, Russia). Extremophiles 12:133–145
Sorokin DY, van Pelt S, Tourov TP, Evtushenko LI (2009) Nitriliruptor alkaliphilus gen. nov., sp. nov., a deep-lineage haloalkaliphilic actinobacterium from soda lakes capable of growth on aliphatic nitriles, and proposal of nitriliruptoraceae fam. nov. and nitriliruptorales ord. nov. Int J Syst Evol Microbiol 59:248–253
Weon HY, Kim BY, Yoo SH, Kim JS, Kwon SW, Go SJ, Stackebrandt E (2006) Loktanella koreensis sp. nov., isolated from sea sand in Korea. Int J Syst Evol Microbiol 56:2199–2202
Yoon JH, Kang SJ, Lee SY, Jung YT, Lee JS, Oh TK (2012) Marivita hallyeonensis sp. nov., isolated from seawater, reclassification of Gaetbulicola byunsanensis as Marivita byunsanensis comb. nov. and emended description of the genus Marivita Hwang et al. Int J Syst Evol Microbiol 62:839–843
WoRMS Editorial Board (2015) World register of marine species. Available from http://www.marinespecies.org at VLIZ
Acknowledgments
This work was supported by research grants CSD 2009–0006 of the Consolider-Ingenio programme and BIO2014-51953-P from the Spanish Ministerio de Ciencia e Innovación, currently the Ministerio de Economía y Competitividad, all including ERDF (European Regional Development Funds). JFAG was supported by a postdoctoral fellowship from CONACyT (Mexico). We wish to thank referees for their constructive commentaries which improved the manuscript.
Conflict of Interest
All authors declare no conflict of interest in this manuscript
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLSX 30 kb)
Figure S1
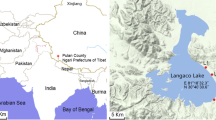
Location of Isabel Island in the Pacific coast of Mexico. Shape of Isabel Island and the crater-lake draw at scale. Sampling sites (1–3) for water and sediment collection are indicated by arrowheads\ (GIF 23 kb)
Figure S2
Bacterial diversity in Isabel soda-lake samples. Rarefaction analysis was based on Operational Taxonomic Units (OTU’s) established at 97 % sequence identity of partial 16S rRNA gene sequences of water and sediment samples from Isabel Island Lake (GIF 12 kb)
Figure S3
16S rRNA gene fingerprint of γ-Proteobacteria isolates from Isabel Island lake samples. Total DNA of selected isolates was digested, electrophoresed and vacuum-blotted onto nylon filters. A PCR fragment (340 bp) of Halomonas venusta (4WSA1) 16S rRNA gene was used as probe. Hybridization fingerprints point to different number of rrn operons (ranging from 2 to 5) according to the genus. Positions of DNA molecular size markers (kbp) are shown on left side (GIF 71 kb)
Rights and permissions
About this article
Cite this article
Aguirre-Garrido, J.F., Ramírez-Saad, H.C., Toro, N. et al. Bacterial Diversity in the Soda Saline Crater Lake from Isabel Island, Mexico. Microb Ecol 71, 68–77 (2016). https://doi.org/10.1007/s00248-015-0676-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-015-0676-6