Abstract
Wolbachia is the most widespread intracellular α-proteobacteria maternally inherited endosymbiont of insects and nematodes. These bacteria are associated with a number of different reproductive phenotypes of their hosts. Relatively few studies have dealt with distribution of infections across populations and with the influence of these bacteria on host genetic diversification and speciation. The aims of this study are to determine the distribution and rate of infection and to characterize the Wolbachia strains associated with Philaenus spumarius spittlebug (Hemiptera) by using multilocus sequencing typing (MLST) analysis and host phylogeography. The results showed that infection rate was significantly different between members of both main mitochondrial phylogenetic lineages of P. spumarius. We detected much higher infection rates of Wolbachia in P. spumarius populations from the north-east clade than the south-west clade. Moreover, the frequency of these infections varied within and outside the contact zone known from the Carpathians. Given the reproductive alterations which are often associated with this endosymbiont, Wolbachia probably maintain genetic differentiation of its hosts in its contact zone in the Carpathians. This is one of the first studies demonstrating the presence of Wolbachia across a large part of the range of insect species, including the contact zone. The spread of Wolbachia in P. spumarius populations can potentially cause speciation by compromising the potential reproductive barrier between infected and uninfected populations. We discuss possible implications of Wolbachia infection inducing cytoplasmic incompatibility in the population dynamics of this spittlebug but confirm that more studies are also required.
Similar content being viewed by others
Introduction
Wolbachia is the most widespread intracellular α-proteobacteria. This maternally inherited endosymbiont is known to infect 15-76 % of insect species [37, 87]. It is found also in many non-insect invertebrates: spiders, mites, crustaceans, and nematodes [29, 37, 64, 65, 89], whereas it is apparently absent in, e.g., mollusks [70]. Wolbachia exists in 13 monophyletic clades: A to N, referred as supergroups [19]; however, supergroups A and B are known to be the most frequent in insects [64, 87]. Wolbachia lives inside the cytoplasm in reproductive tissues and is associated with a number of different reproductive phenotypes in its hosts, such as cytoplasmic incompatibility (CI) [9, 34, 36, 72], feminization, parthenogenesis inducing (PI), male killing, and modifying fecundity [5, 33, 77]. These modifications of the host breed impart a selective advantage, thus allowing Wolbachia to spread efficiently and rapidly into host populations [19, 29]. Furthermore, the ability to manipulate the reproductive properties may have an effect on the host’s biology, ecology, and evolution [4].
Many studies argue that Wolbachia promote rapid speciation by causing reproductive incompatibility between mates, usually from different populations belonging to distinct mitochondrial lineages [10, 62, 77, 82, 94]. The molecular basis of cytoplasmic incompatibility is unknown, but it has been confirmed that the genetic determinants are maternally inherited [62] and correlate with the presence of rickettsia endosymbionts in the arthropods’ gonad tissue (ovaries and testes) [88]. This phenomenon is expressed when an infected male mates with a female that is not infected, when male and female are both infected with two different Wolbachia strains, or when the male is infected with two strains and the female is infected with a single strain of Wolbachia [24]. Examples of CI were found in a diverse range of insects, including flour beetles, Tribolium confusum [61, 84], alfalfa weevils, Hypera postica [31, 46], parasitic wasps, Nasonia [67], planthoppers, Laodelphax striatellus [58, 59], flour moths, Ephestia cautella [13], mosquitoes, Aedes scutellaris [81], and fruit flies, Drosophila simulans [8, 49]. It is also known that Wolbachia may be transmitted horizontally between different hosts (species or members of distant phylogenetic lineages) [88, 90], which can also play a role in speciation.
Wolbachia infections are relatively common in insects although little information is available about the effects of Wolbachia infections in population scale. Populations of insects might not be infected at all: e.g., weevil Centricnemus leucogrammus [40] and Cryptocephalus leaf beetles [51]. On the other hand, some species seemed to be infected across an entire range by the same supergroup and even the same strain: e.g., weevil Polydrusus inustus [41], Chinese rice leafroller Cnaphalocrocis medinalis [15], Indian butterflies Talicada nyseus [2], and leaf beetle Oreina cacaliae [51]. An alternative example concerns species with populations co-infected by different supergroups and/or strains: e.g., grasshopper Chorthippus parallelus [92], bean beetles Callosobruchus chinensis [43], leaf beetle Crioceris quatuordecimpunctata, and Crioceris quinquepunctata [44, 56]. Also, there are known species that some populations infected and others did not: e.g., flower bug Orius strigicollis [86], little fire ant Wasmannia auropunctata [66], and leaf beetle Diabrotica virgifera [24]. In some species, only a portion of individuals are infected within populations: e.g., grasshopper C. parallelus [92]. The latter two examples could concern situations in which all infected populations harbor the same bacteria (single or multiple infection) or each of these populations is infected by another supergroup and/or strain of Wolbachia. A reason for these infection variants could possibly be found in the natural horizontal transmission of Wolbachia by consumption of infected or contaminated food, e.g., plant phloem [18] and/or from parasitoids, e.g., parasitoidal wasps [27, 32]. However, knowledge about infection patterns in populations is relatively poor due to difficulties in studying large numbers of populations and specimens across species ranges and possible diverse compositions of Wolbachia strains in infected populations. Moreover, it is hard to determine if some populations are really uninfected because in some of them, a very low number of individuals could harbor bacteria [89]. Similar problems concern identification of all strains present in some multiple-infected populations, as some strains could infect low numbers of individuals or some strains could dominate others even within a single individual. However, these issues have not been satisfactorily studied thus far.
In recent years, very few studies report cumulative data about Wolbachia infections in populations within contact zones. The paradigmatic work for our studies was performed by Zabal-Aguirre et al. [92], who described two subspecies of the meadow grasshopper C. parallelus that arose in allopatry and next form a secondary contact zone in the Pyrenees. Zabal-Aguirre et al. [92] reported that Wolbachia infection is widespread in C. parallelus populations, where most of the samples featured 76-100 % infected individuals. Moreover, all the analyzed populations of C. parallelus were infected with Wolbachia, although there were differences in infection type. Their studies indicated a significant infection influence on the origin, maintenance, and dynamics of this contact zone. Bella et al. [7] executed consistent studies with C. parallelus. These studies show that Wolbachia may be involved in speciation phenomenon in this grasshopper that is produced by a reproductive barrier. Additional research of three species of the Allonemobius socius complex of crickets from North America in zones of secondary contact including CI caused by the Wolbachia was described by Marshall [52, 53]. Giordano et al. [24] presented the role of Wolbachia bacteria in reproductive incompatibilities and hybrid zones of Diabrotica beetles and Gryllus crickets. The examples mentioned above suggest that the role of Wolbachia in inducing CI and the resulting speciation in insects could be underestimated. However, most of this research was executed on orthopteran, which is due to intensive studies on this group of insects rather than special association of Wolbachia with these insects. On the other hand, almost nothing is known about the influence of Wolbachia on reproductive barriers, contact zone origin, and speciation process in other groups of insects. Recently, the contact zone has been described for the spittlebug Philaenus spumarius (L.) (Hemiptera: Aphrophoridae) [48, 55], which has been one of the most intensively studied bugs in recent years [20, 21, 25, 48, 54, 55, 68, 71], making it a model species in evolutionary studies of insects.
Currently, genus Philaenus is believed to consist of nine or ten species [20, 54, 80]. Most of them are distributed in the Mediterranean area. Only P. spumarius is widespread and occurs naturally throughout the entire temperate and warm Holarctic region [20, 21]. It therefore seems to be an excellent subject of research for the spread of Wolbachia infection. Recent studies on the phylogeny and population genetics of spittlebugs of the genus Philaenus show that P. spumarius is divided into two highly distinct mitochondrial clades: northeastern (north-central Europe and Asia) and southwestern (Western Europe and the Mediterranean region, also introduced/invaded populations to the North America), which meet along European mountain ranges [48, 54, 55]. Similarly, analysis of a nuclear marker (elongation factor 1 alpha gene (EF1-α)) suggests that there are three main clades: northeastern (Eurasian), southeastern (east Mediterranean-Caucasian), and southwestern (Italo-Iberian), which probably overlap along European mountain ranges [48, 55]. Moreover, detailed examination of genetics for populations in the contact zone of main phylogenetic lineages in the Carpathians demonstrated that some of these populations consist of individuals belonging to different clades and, even more, that some individuals showed signs of hybrid genotypes, including examples of heteroplasmy [48]. These findings strongly suggest that P. spumarius is a complex of evolutionary units with uncertain taxonomic status. Therefore, it is an excellent subject for further population genetic and speciation studies. Because our preliminary analyses confirmed that some of P. spumarius populations are infected by Wolbachia, we decided to perform detailed studies on these bacteria occurrence across the range of P. spumarius, including populations in contact zones.
The principal aims of this study were to determine infection diffusing of Wolbachia and identify supergroups of this microorganism in P. spumarius populations in the entire range and in the contact zone of the main phylogenetic lineages. As a result, we will try to verify the following hypotheses: (i) Wolbachia infects only one of P. spumarius mitochondrial phylogenetic lineages and consequently (ii) Wolbachia infection is associated with the beginning of allopatric speciation of the host and with limited hybridization between genetically distinct populations of the P. spumarius. As this work is the first step in understanding interactions between Wolbachia and P. spumarius, it does not include examination of cytoplasmic incompatibility or other mechanisms influencing reproduction of the host.
Materials and Methods
Sampling Area
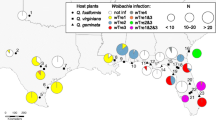
Individuals from 49 populations of P. spumarius were collected from 2003 to 2011 (Fig. 1). Thirty-one populations (102 individuals) covering nearly the entire range of P. spumarius (locality symbols S) and eighteen populations (72 individuals) from six transects across the Carpathian arc (locality symbol CM) were sampled (Table 1). Additionally, six specimens of Philaenus tesselatus (from Portugal) and single representatives of Philaenus italosignus (from Sicily), Philaenus signatus (from Greece), Philaenus arslani (from Lebanon), Philaenus loukasi (from Greece), Philaenus tarifa, and Philaenus maghresignus (from Southern Spain) were tested for Wolbachia presence (we missed the newly described species Philaenus elbursianus and Philaenus iranicus due to the unavailability of these Iranian taxa [80]). Samples of these species, as well as the majority of individuals of P. spumarius, were previously used in phylogenetic and phylogeographic studies [54, 55], whereas Carpathian samples were used for examination of the contact zone [48]. The spittlebugs were caught in a sweep-net, instantaneously preserved in 99 % ethanol, and stored at −20 °C. All tested specimens were damaged during extraction of DNA procedure. The remaining voucher specimens are preserved at the Institute of Systematics and Evolution of Animals, Polish Academy of Sciences.
Localization of P. spumarius sampling sites and distribution of Wolbachia infections (including Carpathian Mountains (CM)). Circles represent populations harboring haplotypes belonging to SW mitochondrial clade; triangles represent populations with NE mitochondrial haplotypes; and squares represent populations including haplotypes from both clades. Empty marks indicate lack of evidence for Wolbachia infection; grey marks represent populations in which infection was detected (strains belonging to B supergroup). Populations infected with local strain of A supergroup are marked additionally with a plus mark
Wolbachia Detection
DNA was extracted from the whole body of the individuals. Amplification, purification, and sequencing of multilocus sequence typing (MLST) genes were performed using the standard protocols with different sets of primers (available at: http://www.pubmlst.org/wolbachia/) [6]. A total of 174 specimens of P. spumarius and 12 specimens of other Philaenus species were screened for the presence of Wolbachia strains. Detection was based on the Wolbachia ftsZ gene and results in the amplification of an approximately 528-base pair-long DNA fragment with the Wolbachia primers ftsZ_F and ftsZ_R [6]. Next, all the ftsZ-positive specimens were subjected to amplification for the other MLST genes (gatB, coxA, hcpA, and fbpA). To rule out the possibility of poor quality or degraded DNA templates used for amplification, the cytochrome B gene of mitochondrial DNA (CB-J10747 and CB-N11526 primers by Stewart i Beckenbach [74]) was additionally amplified. Polymerase chain reaction (PCR) products were checked on agarose gel. Sequences of the same gene (not generated de novo but obtained from previous studies, Maryańska-Nadachowska et al. [48, 54, 55] were used in further analyses.
PCR was performed in 30 μl reaction volumes with 3.0 μl of 10× PCR buffer, 3.0 μl of 25 mM MgCl2, 0.6 μl of a dNTP mixture, each in a 10mM concentration, 0.6 μl of each 15 mM forward and reverse primers, 3.0 μl of 100 ng of genomic DNA, 0.2 μl of Taq DNA polymerase (Qiagen, Germany) and sterile and deionized water (up to 30.0 μl). PCR conditions for standard primers were as follows: 4 min at 95 °C followed by 35 cycles of 30 s at 95 °C, 1 min at 54 °C, and 2 min at 72 °C, with 10 min at 72 °C after the last cycle. To obtain a full set of MLST sequences for individuals infected with local strain (found in specimens located in the inner side of the southern Carpathians, see “Results”), some genes need to be amplified by specific primers, with the annealing temperature adjusted respectively from protocols available at: http://www.pubmlst.org/wolbachia/. PCR products were subjected to electrophoresis on 1.5 % agarose gels and stained with Midori Green DNA Stain from ABO. For the MLST gene sequencing of infected P. spumarius, specimens were obtained with the same primers used during amplification (available at http://www.pubmlst.org/wolbachia/).
MLST Analyses of Wolbachia Strains
The sequences were compared with the online NCBI databank using the Basic Local Alignment Search Toot (BLAST) [1] option to check if primers specifically amplified the targeted α-proteobacteria. Next, sequences were edited using the BioEdit Sequence Alignment Editor 5.0.9. [26] and aligned using ClustalX 1.8 [79]. Haplotype (allele) reconstruction of MLST genes from genotype data was conducted using the algorithms provided in PHASE as implemented in DnaSP 5.0 [47].
The information about host and Wolbachia haplotypes has been submitted to the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/BankIt/). The nucleotide sequences have been deposited in the GenBank database at accession numbers for hcpA, KM377676–KM377688; coxA, KM377689–KM377736; fbpA, KM377737–KM377764; ftsZ, KM377765–KM377773; and gatB, KM377774–KM377832.
The Wolbachia strains were assigned a sequence type (ST) and defined as the combination of five integers corresponding to the allele numbers at the five MLST loci (allelic profile) with the help of START2 software [38]. A strain is defined as a Wolbachia isolate from a single host population. Strains identical to the five alleles were assigned the same ST.
Sequences of MLST genes were compared against Wolbachia sequences deposited in GenBank (which used BLAST) to find other hosts harboring the most similar (maximum identity) Wolbachia strains sequences. A dataset of the five concatenated Wolbachia MLST gene (i.e., gatB, coxA, hcpA, fbpA, and ftsZ), belonging to 87 different Wolbachia STs were retrieved from the MLST Website and used in comparative analysis with the STs isolated from P. spumarius.
Statistical Analyses
The differences of frequencies of positive and negative spittlebug for Wolbachia from the whole species range, contact zone in the Carpathians, and all populations outside this zone were analyzed by using the Chi-square test. Relationships between Wolbachia strains were featured by building phylogenetic trees and networks for MLST genes. Trees and networks were made of original sequences for each MLST gene separately and for a combined dataset of all MLST sequences. Phylogenetic trees were constructed using MEGA5.0 software [78] with neighbor-joining algorithm. Bootstrap analysis was done with 1000 replications; bootstrap values were calculated using a 50 % majority rule. Construction of networks was done in SplitsTree4 [35]. This software use of median-joining algorithm distance estimates to compute unrooted phylogenetic networks from molecular sequence data. Contrary to traditional phylogenetic trees, it allows for visualization of multiple connections among examined sequences which could represent, e.g., recombination events. Moreover, mitochondrial DNA (mtDNA) networks from previous phylogeographic studies on P. spumarius [55, 48] were used for visualization connections with Wolbachia infections against a background of P. spumarius main phylogenetic lineages. The mtDNA network was build with the use of TCS 1.21 software [17], and symbols of haplotypes (circles) were colored according to Wolbachia infection prevalence (lack of infection, single or all individuals infected). The same relationship was visualized on the mitochondrial phylogeographic map of species range. The pairwise homoplasy test (PHI) [14] has been shown to identify the presence/absence of recombination within a wide range of sequence samples with a low false-positive rate [14]. PHI test was used to analyze genetic recombination within and among MLST genes. Results were obtained in part by using SplitsTree4 [35].
Results
Wolbachia Detection in Philaenus Species
The screening of Wolbachia infections among eight Philaenus species was initially executed using amplification of ftsZ gene. Wolbachia infections were found in P. spumarius and P. italosignus specimens. However, in the other Philaenus species - P. signatus, P. arslani, P. loukasi, P. tarifa, P. maghresignus, and P. tesselatus - we did not ascertain infections of these bacteria. However, it is important to highlight that only for P. spumarius did we screen many samples of specimens across species range; for the rest of species, we tested only single specimens for Wolbachia infections.
Infection Rate of Wolbachia in Different P. spumarius Populations
In 23 different P. spumarius populations, we obtained an approximately 528-base pair-long DNA fragment out of the ftsZ gene for 72 of 174 individuals. This indicates that 41.4 % of the individuals from all examined populations were positive for infection (Table 2).
Additionally obtained results clearly show differences in infections among populations and main mitochondrial clades. When considering all sampled populations, 70.3 % of individuals from the NE clade were found to be infected, whereas only 20.0 % of the individuals from the SW possessed these bacteria, respectively (Table 2). But these infection frequencies varied within and outside contact zone.
In the identified contact zone in the Carpathians, 93.9 % individuals belonging to the NE clades were infected but two times fewer (46.2 %) individuals from the SW clade were Wolbachia positive (Table 2). In total, 68.1 % of all Carpathian specimens were positive for Wolbachia infections.
In the rest of species range (excluding Carpathians), there were in total 51.2 % of infected specimens from the NE clade; only 3.3 % of individuals from the SW clade possessed these bacteria (Table 2).
The infection rate was significantly different between members of both main mitochondrial phylogenetic lineages when considering the whole range of species (χ 2 = 45.5, p < 0.0001), contact zone in the Carpathians (χ 2 = 58.4, p < 0.0001), and all populations outside this zone (χ 2 = 50.3, p < 0.0001).
Distribution of Wolbachia in P. spumarius
The sequence analyses of all MLST genes indicates that supergroup B occurred in most of the infected populations in the range of P. spumarius, although there were differences in infection type and variations of strains.
In the group of all infected specimens, nearly 75 % harbored more than one strain of supergroup B, in the Carpathians, and over 76 % were double infected. Most individuals were from the eastern Carpathians (from the outer and central arch of the mountains) (Fig. 1).
In the rest of the range, 36 % were single infected by a strain that was not defined in the MLST database. They occurred mostly in populations where single individuals were infected (Portugal-S4, France-S5, U.S.A.-S15, and Georgia-S21) (Fig. 1).
Throughout the Carpathian’s contact zone, most specimens were infected by the same Wolbachia supergroup like other infected populations from Europe and Asia; moreover, there were only four populations without Wolbachia infections, all from the southwestern clade (Fig. 1). In three closely located populations (CM-59, CM-65, and CM-67) from southern Carpathians (Fig. 1), unique strains of these bacteria were identified as belonging to supergroup A. Of all of the obtained, only approximately 5 % of all infections were from a distinct supergroup, and all were from the inner side of the southern Carpathians (Fig. 1). This different supergroup has been detected in all infected individuals from CM-65 and CM-67 belonging to the southwestern mitochondrial clade and one specimen from the population CM-59 belonging to the northeastern clade. In the CM-59 population, only one of four specimens harbored this local strain; three other infected specimens harbored similar strains like other multi-infected individuals from SW. Moreover, three belonging to supergroup A/B populations were single infected with a distinct strain (not detected in MLST database). Only the CM-67 specimen was double infected with different strains of this supergroup. Moreover, analysis of MLST allelic profiles (Table 1) indicated only two identical sequence types from the 87 obtained. This testified to the very high variability of the Wolbachia-analyzed genes.
Additionally, there were identified hosts harboring similar strains of Wolbachia as P. spumarius, obtained from the GenBank (Table 2). The strains of this bacterium in P. spumarius were closely related to those in wasps (Vespidae), flies (Drosophilidae), hemipterans (Aleyrodidae), and also in beetles (Chrysomelidae, Curculionidae).
Phylogenetic Analysis of the Wolbachia MLST Sequences
The phylogenetic analysis results of the Wolbachia MLST sequences from P. spumarius and P. italosignus are shown in Fig. 2. The neighbor-joining analysis revealed two major branches in the phylogenetic trees based on Wolbachia MLST sequences separately for each gene (Fig. S1a-e) and collectively for all MLST genes (Fig. 2). These two characteristic branches clustered Wolbachia sequences from the P. spumarius populations into two main supergroups (Fig. 2). At the first branches are nearly 95 % of populations which harbored strains belonging to the B supergroup. The second branch included Wolbachia-infected populations from the southern Carpathians, which suggested that approximately 5 % were infected from an alternative source by strains belonging to supergroup A. Additionally, the possible relationships between the Wolbachia haplotypes were estimated using the median-joining networks shown in Figs. 3 and S2a-e, which show congruent patterns with neighbor-joining trees. Additionally this network brought more information than traditional phylogenetic tree as it showed also multiple connections among examined Wolbachia haplotypes (MLST strains) which could correspond to, e.g., recombination events.
Moreover, the networks in Fig. 4 demonstrated results of relations within the P. spumarius mtDNA clades and infections of Wolbachia bacteria. This figure clearly shows that infections dominate in northeastern clade, and only in population from Norway (belonging to this clade) we did not find this microorganism. In the southwestern clade, the mtDNA network the most infected populations were from the Carpathian Mountains where the contact zone of the main phylogenetic P. spumarius clades occurs. This is where specimens can mix with the northeastern clade. Another single exception of the infected populations belonging to the southwestern clade is from Portugal and the USA. About one fifth of the infected individuals was in the population from Portugal; in the USA, one-third of the examined individuals harbored the Wolbachia.
Haplotype cytochrome B networks of P. spumarius and association of Wolbachia infections with haplotypes belonging to the NE and SW mitochondrial clades from the species range, including contact zone in the Carpathians (symbols of populations like in Table 1). White circles, not infected; gray circles, all population infected; black circle, some individuals infected of population
Recombination Analysis
The phi test executed by SplitsTree4 [35] indicated statistically significant evidence for recombination of three of five analyzed MLST genes: gatB, coxA, and fbpA but not for hcpA and ftsZ (Table 3). Moreover, recombination was detected among joined sequences of all MLST genes (Table 3).
Discussion
This study provides the first evidence of Wolbachia infections in Philaenus spittlebugs; moreover, it represents primary research that includes distribution of these bacteria among P. spumarius populations across almost the entire species range and in the contact zone of the main phylogenetic lineages of this insect in the Carpathians.
Since first discovered in Culex pipiens [28], Wolbachia have been described as a widespread and common bacteria infecting insects all over the world (e.g., Neotropics [83], Palaearctics [91], and Nearctics [89]). Most research on Wolbachia focused on screens of species, but single representatives of each species were usually investigated. Due to known effects of Wolbachia on reproduction of its hosts (e.g., cytoplasmic incompatibility (CI) [10, 11, 45, 62–64], parthenogenesis [3, 75, 76], male killing [23, 33] and feminizing of genetic males [30, 69]), it can be hypothesized that these bacteria have influenced on diversification and speciation of its hosts. Unfortunately, such studies are not easy to perform due to problems with identifying contact zones of species/phylogenetic lineages that harbor distinct strains or when only one species/phylogenetic lineage is infected. Consequently, there were only single studies dealing with infections in the contact zone. Example of such recent research might be work done by Zabal-Aguirre et al. [92, 93], which shows the genetic structure of Wolbachia infection in the Pyrenean contact zone of the widely investigated Meadow grasshopper C. parallelus subspecies: European C. p. parallelus and C. p. erythropus.
In this study, we discovered that approximately 41 % specimens of P. spumarius spittlebug across its entire range are Wolbachia positive, but the infection rate differs significantly between members of mitochondrial clades (c. 51 % infected in NE clades versus only c. 3 % in the SW clade) and in contact zone (c. 93 % from the NE clade and c. 46 % from the SW clade in this zone). These frequencies support the idea that Wolbachia is widespread in P. spumarius just like in numerous other arthropods [19, 29, 37, 64, 87], but that these bacteria are not distributed uniformly. Moreover, the higher frequency of infections in the contact zone compared with other parts of the species range, especially noticeable for the SW clade, indicates that Wolbachia could spread between members of both mitochondrial clades in areas where they meet. This implies that mitochondrial phylogenetic lineages of P. spumarius have not yet formed sufficient reproductive barriers, which has also recently been proven on the basis of population genetic studies [48]. It is probable that some environmental factors (basically, host plants that can mediate Wolbachia infection in phytophagous insect populations [12, 16, 85]), could be responsible for the observed pattern of these bacteria distribution and diversity in P. spumarius range. P. spumarius is a polyphagous species, but it is possible that its populations from NE and SW could feed on different plant species due to differences in vegetation between the temperate flora of lowland central and eastern Europe and north Asia compared with Mediterranean flora of southern Europe (where most Philaenus species feed exclusively on Asphodelus) [50]. The contact zone in the Carpathians simultaneously forms a potential primary contact zone for Wolbachia diffusion into the SW clade. Outside this zone, an almost strict division was observed: infection in most populations belonging to the NE clade and almost no infection in populations belonging to the SW clade.
The increased level of infection in P. spumarius NE clade and only isolated infections of these bacteria in the SW clade indicates that the ancestors of the lineage were presumably infected by Wolbachia after the split of its main mitochondrial lineages or during this split (which possibly has not finished yet). The distribution of Wolbachia could also be associated with the presence of some host plants in the P. spumarius range (see explanation above). The demographic and spatial expansion of this species may have occurred earlier than Holocene [55, 68]. Also, high genetic diversity detected in P. spumarius populations from northern Europe (Scandinavia) indicate that the north of Europe was colonized by populations that may have survived in several extra-Mediterranean glacial refugia in addition to the “classical” Mediterranean refuges [68]. That may explain non-infected populations from Norway; however, loss of infection in these populations cannot be ruled out. Also interesting are the non-infected populations from Crimea and southern Russia (belonging to distinct lineage within the SW clade), which apparently avoided infection despite close proximity to populations belonging to the infected NE clade. The possible source of the infection is directed to the eastern locations (possibly from Asia) and derived from there to the west, particularly to central Europe but not to northern and southeastern Europe. This infection wave has not invaded southern populations. However, single infections in southwestern P. spumarius populations from Portugal and probably descendent population in the USA (according to mitochondrial studies [55]) can be explained by accidental human translocations. Moreover, lack of infection in southern populations of P. spumarius is consistent with lack of infection in other Philaenus species that inhabit mostly Mediterranean regions. The single infection in P. italosignus might have come from feeding on plant phloem [18], which could have mediated Wolbachia transmission from P. spumarius or other infected insect.
It is probable that the observed pattern of Wolbacha infection in the contact zone may have caused cytoplasmic incompatibility between isolated populations from the northeast and the southwest. In this study, we have a few examples of populations where individuals from two main mitochondrial clades were in one population collectively, and infections were detected only in specimens from NE clades. That support of cytoplasmic incompatibility may have also occurred between infected and non-infected P. spumarius individuals. Therefore, this phenomenon may also play a role with allopatric speciation in P. spumarius. This phenomenon needs further studies, including mating of hosts belonging to distinct mitochondrial lineages and investigation of Wolbacha transmission to future generations. Moreover, this phenomenon should also be tested with respect to feeding preferences of P. spumarius, which are only broadly known as polyphagous species, but no detailed studies have been undertaken to verify differences in host plant composition across its range, e.g., with the use of host plant barcoding from insect guts (e.g., [42, 57]). Wolbachia could be transmitted by means of plant phloem, so this route of infection should also be considered in further studies on P. spumarius.
As was described in the “Results,” different infection types show that P. spumarius were infected by more than one supergroup of Wolbachia. Moreover, different strains were detected in both B and A supergroups, which feature advanced states of infection of this microorganism and a high degree of recombination events. Supergroup B is a main group of Wolbachia in P. spumarius populations, but local infections of supergroup A in the southern Carpathians showed that infections is not homogeneous in all populations. That state can be explained by different sources of infections (e.g., from other insect species) that occurred only locally in southern Carpathians and have not spread to other populations of P. spumarius. This local distribution of supergroup A could additionally be explained by the cytoplasmic incompatibility of individuals harboring this supergroup with bugs infected (or not infected) by supergroup B. As we show in Table 2, P. spumarius would have received Wolbachia from other insect species. The main strain of these bacteria probably might come from wasps (Vespidae), flies (Drosophilidae), or hemipterans (Aleyrodidae), so infection could be transmitted either by predation (some injury, e.g., by wasp) or more probably by parasitoids, which may function as a vector for Wolbachia bacteria and transfer it to other arthropods [39, 60]. Instead of supergroup A in the local area in the Carpathians, beetles (Chrysomelidae, Curculionidae) were identified as the most presumable donor of Wolbachia.
In this study, we acknowledge our main hypothesis that Wolbachia infects in the majority the NE mitochondrial clade of P. spumarius. Our results pointed out single infections in SW clade, which could have been caused by man-made translocation of these bugs. Although, individuals from the Carpathians belonging to the SW clade harbored different supergroup of Wolbachia. There is very likely that between two main mitochondrial P. spumarius clades, we are dealing with cytoplasmic incompatibility. The presented results indicate that Wolbachia may play a significant role in the maintenance of the limited hybridization between genetically distinct populations of the P. spumarius in the contact zone via probable cytoplasmic incompatibility. This phenomenon in the Carpathians should be considered to probably exist in the other P. spumarius contact zones such as in the Alps, the Pyrenees and the Caucasus. We also verified the correlation of mitochondrial variability with Wolbachia infection through this contact zone. Wolbachia are associated with mtDNA, as both are maternally inherited within cytoplasm in reproductive tissue and can therefore result in a homogenization of mtDNA haplotypes (selective slippage [22, 72, 73]). Consequently, this would have implications for important evolutionary processes along with speciation. There was evidence presented in other research of Nasonia of reproductive isolation among species driven by Wolbachia [92]. Our results clearly show the distribution of Wolbachia in the P. spumarius range and reveal geographical patterns of distribution of the bacterial strains that may also appear in other organisms. But our results do not represent complete evidence that Wolbachia acts as a barrier to reproduction in the contact zone of P. spumarius by inducting cytoplasmic incompatibility, although overall they give a view that is consistent with such a possibility.
Conclusions
This work presents the hypothesis that Wolbachia infection is associated with limited hybridization between genetically distinct populations of the P. spumarius and also with allopatric speciation in progress in the secondary contact zone in the Carpathians. Yet, further study is also required for (i) infections in other contact zones in the Alps, the Pyrenees, and the Caucasus, (ii) laboratory crossing of individuals from different populations (mtDNA and infection) to detect signs of cytoplasmic incompatibility and reinforcement of reproductive barriers, and (iii) artificial trials of infection of individuals from uninfected populations and removal of bacteria from infected populations to check influence of such experiments on host survival and reproduction. This data overall supports the hypothesis that Wolbachia infections are associated with only one P. spumarius clade. Overall, this suggests ongoing partial cytoplasmic incompatibility in the hybridization events among the P. spumarius clades in the Carpathians and an allopatric speciation in progress.
References
Altschul S, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ankola K, Brueckner D, Puttaraju HP (2011) Wolbachia endosymbiont infection in two Indian butterflies and female-biased sex ratio in the Red Pierrot Talicada nyseus. J Biosci 36:845–850
Arakaki N, Miyoshi T, Noda H (2001) Wolbachia-mediated parthenogenesis in the predatory thrips Franklinothrips vespiformis (Thysanoptera: Insecta). Proc R Soc Lond B 268:1011–1016
Augustinos AA, Santos-Garcia D, Dionyssopoulou E, Moreira M, Papapanagiotou A et al (2011) Detection and characterization of Wolbachia infections in natural populations of aphids: is the Hidden Diversity Fully Unraveled? PLoS ONE 6:12
Baldo L, Lo N, Werren JH (2005) Mosaic nature of the Wolbachia surface protein. J Bacteriol 187:5406–5418
Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, Hayashi C, Maiden MCJ, Tettelin H, Werren JH (2006) Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol 72:7098–7110
Bella JL, Martínez-Rodríguez P, Arroyo-Yebras F, Bernal A, Sarasa J, Fernández-Calvín B et al (2010) Wolbachia infection in the Chorthippus parallelus hybrid zone: evidence for its role as a reproductive barrier. J Orthop Res 19:205–212
Binnington KL, Hoffmann AA (1989) Wolbachia-like organisms and cytoplasmic incompatibility in Drosophila simulans. J Invertebr Pathol 54:344–352
Bordenstein SR, Wernegreen JJ (2004) Bacteriophage Xux in endosymbionts (Wolbachia): infection frequency lateral transfer and recombination rates. Mol Biol Evol 21:1981–1991
Breeuwer JAJ, Werren JH (1990) Microorganisms associated with chromosome destruction and reproductive isolation between two insect species. Nature (London) 346:558–560
Breeuwer JAJ, Stouthamer R, Barns SM, Pelletier DA, Weisburg WG, Werren JH (1992) Phylogeny of cytoplasmic incompatibility microorganisms in the parasitoid wasp genus Nasonia (Hymenoptera: Pteromalidae) based on 16S ribosomal DNA sequences. Insect Mol Biol 1:25–36
Broderick NA, Goodman RM, Handelsman J, Raffa KF (2003) Effect of host diet and insect source on synergy of gypsy moth (Lepidoptera: Lymantriidae) mortality to Bacillus thuringiensis subsp kurstaki by zwittermicin A. Environ Entomol 32:387–391
Brower JH (1975) Cytoplasmic incompatibility: occurrence in a stored-product pest Ephestia cautella. Ann Entomol Soc Am 69:1011–1015
Bruen TC, Philippe H, Bryant D (2006) A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681
Chai HN, Du YZ, Qiu BL, Zhai BP (2011) Detection and phylogenetic analysis of Wolbachia in the Asiatic rice leafroller Cnaphalocrocis medinalis in Chinese populations. J Insect Sci 11:123
Charlat S, Hurst GDD, Merçot H (2003) Evolutionary consequences of Wolbachia infections. Trends Genet 19:217–223
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
DeLay BD (2013) Symbionts associated with the salivary glands of the potato leafhopper Empoasca fabae and their function when feeding on leguminous hoss. Digital Repository at the University of Maryland University of Maryland (College Park Md.)
Doudoumis V, Tsiamis G, Wamwiri F, Brelsfoard C, Alam U, Aksoy E et al (2012) Detection and characterization of Wolbachia infections in laboratory and natural populations of different species of tsetse flies (genus Glossina). BMC Microbiol 12(Suppl 1):S3
Drosopoulos S (2003) New data on the nature and origin of colour polymorphism in the spittlebug genus Philaenus (Hemiptera: Aphrophoridea). Ann Soc Entomol Fr 39:31–42
Drosopoulos S, Maryańska-Nadachowska A, Kuznetsova VG (2010) The Mediterranean: area of origin of polymorphism and speciation in the spittlebug Philaenus (Hemiptera Aphrophoridae). Zoosyst Evol 86:125–128
Dumas E, Atyame CM, Milesi P, Fonseca DM, Shaikevich EV, Unal S et al (2013) Population structure of Wolbachia and cytoplasmic introgression in a complex of mosquito species. BMC Evol Biol 13:181
Fialho RF, Stevens L (1997) Molecular evidence for single Wolbachia infections among geographic strains of the flour beetle Tribolium confusum. Proc R Soc Lond B 264:1065–1068
Giordano R, Jackson JJ, Robertson HM (1997) The role of Wolbachia bacteria in reproductive incompatibilities and hybrid zones of Diabrotica beetles and Gryllus crickets. Proc Natl Acad Sci USA Evol 94:11439–11444
Halkka O, Halkka L (1990) Population genetics of the polymorphic spittlebug Philaenus spumarius (L). Evol Biol 24:149–191
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows95/98/NT. Nucleic Acid Symp Ser Lond 41:95–98
Heath BD, Butcher RDJ, Whitfield GF, Hubbard SF (1999) Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr Biol 9:313–316
Hertig M (1936) The rickettsia Wolbachia pipientis (gen. et sp. n.) and associated inclusions of the mosquito Culex pipiens. Parasitology 28:453–486
Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH (2008) How many species are infected with Wolbachia? - a statistical analysis of current data. FEMS Microbiol Lett 281:215–222
Hiroki M, Kato Y, Kamito T, Miura K (2002) Feminization of genetic males by a symbiotic bacterium in a butterfly Eurema hecabe (Lepidoptera: Pieridae). Naturwissenschaften 89:167–170
Hsiao C, Hsiao TH (1985) Rickettsia as the cause of cytoplasmic incompatibility in the alfalfa weevil Hypera postica. J Invertebr Pathol 45:244–246
Huigens ME, de Almeida RP, Boons PAH, Luck RF, Stouthamer R (2004) Natural interspecific and intraspecific horizontal transfer of parthenogenesis-inducing Wolbachia in Trichogramma wasps. Proc R Soc Lond B Biol Sci 271:509–515
Hurst GDD, Jiggins FM, Von der Schulenburg JHG, Bertrand D, West SA, Goriacheva II et al (1999) Male killing Wolbachia in two species of insect. Proc R Soc Lond B 266:735–740
Hurst GDD, Werren JH (2001) The role of selfish genetic elements in eukaryotic evolution. Nat Rev Genet 2:597–606
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Jaenike J, Dyer KA, Cornish C, Minhas MS (2006) Asymmetrical reinforcement and Wolbachia infection in Drosophila. PLoS Biol 4:e325
Jeyaprakash A, Hoy MA (2000) Long PCR improves Wolachia DNA amplification: wsp sequences found in 76 % of sixty-three arthropod species. Insect Mol Biol 9:393–405
Jolley KA, Feil EJ, Chan MS, Maiden MC (2001) Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230–1231
Kacsoh BZ, Lynch ZR, Mortimer NT, Schlenke TA (2013) Fruit flies medicate offspring after seeing parasites. Science 339:947–950
Kajtoch Ł (2011) Conservation genetics of xerothermic beetles in Europe: the case of Centricnemus leucogrammus. J Insect Conserv 15:787–797
Kajtoch Ł, Korotyaev B, Lachowska-Cierlik D (2012) Genetic distinctness of parthenogenetic forms of European Polydrusus weevils of the subgenus Scythodrusus. Insect Sci 19:183–194
Kajtoch Ł (2014) A DNA metabarcoding study of a polyphagous beetle dietary diversity: the utility of barcodes and sequencing techniques. Folia Biol 2:223–234
Kondo NI, Tuda M, Toquenaga Y, Lan YC, Buranapanichpan S, Horng SB et al (2011) Wolbachia infections in world populations of bean beetles (Coleoptera: Chrysomelidae: Bruchinae) infesting cultivated and wild legumes. Zool Sci 28(7):501–508
Kubisz D, Kajtoch Ł, Mazur MA, Lis A, Holecová M (2012) Conservation genetics of highly isolated populations of the xerothermic beetle Criocerisquatuordec impunctata (Chrysomelidae). Invertebr Biol 131:333–344
Laven H (1967) Speciation and evolution in Culex pipiens. In: Wright J, Pal R (eds) Genetics of insect vectors of disease. Elsevier, Amsterdam, pp 251–275
Leu SJC, Li JKK, Hsiao TH (1989) Characterization of Wolbachia postica the cause of reproductive incompatibility among alfalfa weevil strains. J Invertebr Pathol 54:248–259
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lis A, Maryańska-Nadachowska A, Lachowska-Cierlik D, Kajtoch Ł (2015) The secondary contact zone of phylogenetic lineages of the spittlebug Philaenus spumarius: an example of incomplete allopatric speciation? J Insect Sci 14(227). doi:10.1093/jisesa/ieu089
Louis C, Nigro L (1989) Ultrastructural evidence of Wolbachia Rickettsiales in Drosophila simulans and their relationships with unidirectional cross in compatibility. J Invertebr Pathol 54:39–44
Loukas M, Drosopoulos S (1992) Population genetics of the spittlebug genus Philaenus (Homoptera: Cercopidae) in Greece. Biol J Linn Soc 46:403–413
Montagna M, Gómez-Zurita J, Giorgi A, Epis S, Lozzia G, Bandi C (2014) Metamicrobiomics in herbivore beetles of the genus Cryptocephalus (Chrysomelidae): toward the understanding of ecological determinants in insect symbiosis. Insect Sci. doi:10.1111/1744-7917.12143
Marshall JL (2004) Allonemobius-Wolbachia host-endosymbiont system: evidence for rapid speciation and against reproductive isolation driven by cytoplasmic incompatibility. Evolution 58:2409–2425
Marshall JL (2007) Rapid evolution of spermathecal duct length in the Allonemobius socius complex of crickets: species population and Wolbachia effects. PLoS ONE 2:e720
Maryańska-Nadachowska A, Drosopoulos S, Lachowska D, Kajtoch Ł, Kuznetsova VG (2010) Molecular phylogeny of the Mediterranean species of Philaenus (Hemiptera: Auchenorrhyncha: Aphrophoridae) using mitochondrial and nuclear DNA sequences. Syst Entomol 35:318–328
Maryańska-Nadachowska A, Kajtoch Ł, Lachowska D (2012) Genetic diversity of Philaenus spumarius and P. tesselatus (Hemiptera: Aphrophoridae): implications for evolution and taxonomy. Syst Entomol 37:55–64
Mazur MA, Olbrycht T, Szewkienicz A (2014) Magdalis (Odontomagdalis) caucasica (Tournier 1872) (Coleoptera: Curculinoidae) - a first record in Poland and summary data on the occurrence in Europe. Balt J Coleopterol 14:53–58
Navarro SP, Jurado-Rivera JA, Gómez-Zurita J, Lyal CHC, Vogler AP (2010) DNA profiling of host herbivore interactions in tropical forests. Ecol Entomol 35:18–32
Noda H (1984) Cytoplasmic incompatibility in allopatric field populations of the small brown planthopper Laodelphax striatellus in Japan. Entomol Exp Appl 35:263–267
Noda H (1987) Further studies on cytoplasmic incompatibility in local populations of Laodelphax striatellus in Japan (Homoptera: Delphacidae). Appl Entomol Zool 22:443–448
Oliver KM, Russell JA, Moran NA, Hunter MS (2003) Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci U S A 100:1803–1807
O’Neill SI (1989) Cytoplasmic symbionts in Tribolium confusum. J Invert Pathol 53:132–134
O’Neill SO, Karr TL (1990) Bidirectional cytoplasmic incompatibility between conspecific populations of Drosophila simulans. Nature 348:178–180
O’Neill SL, Giordano R, Colbert AME, Karr TL, Robertson HM (1992) 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci U S A 89:2699–2702
Prakash BM, Puttaraju HP (2007) Frequency of infection with A and B super group Wolbachia in insects and pests associated with mulberry and silkworm. J Biosci 32:671–676
Puttaraju HP, Madhu M (2002) Presence of Wolbachia endosymbionts in different silkworm species and races and in their uzifly parasites. J Invertebrate Pathol 79:120–122
Rey O, Estoup A, Facon B, Loiseau A, Aebi A et al (2013) Distribution of endosymbiotic reproductive manipulators reflects invasion process and not reproductive system polymorphism in the little fire ant Wasmannia auropunctata. PLoS ONE 8:e58467
Richardson PM, Holmes WP, Saul GBII (1987) The effect of tetracycline on reciprocal cross incompatibility in Mormoniella [D. Nasonia] vitripennis. J Invertebr Pathol 50:176–183
Rodrigues ASB, Silva SE, Marabuto E, Silva DN, Wilson MR et al (2014) New mitochondrial and nuclear evidences support recent demographic expansion and an atypical phylogeographic pattern in the spittlebug Philaenus spumarius (Hemiptera: Aphrophoridae). PLoS ONE 9:e98375
Rousset F, Bouchon D, Pintureau B, Juchault P, Soligna M (1992) Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods. Proc R Soc Lond B 250:91–98
Schilthuizen M, Gittenberger E (1998) Screening mollusks for Wolbachia infection. J Invertebr Pathol 71:268–270
Seabra SG, Pina-Martin SF, Marabuto E, Yurtsever S, Halkka O, Quartau JA, Paulo OS (2010) Molecular phylogeny and DNA barcoding in the meadow-spittlebug Philaenus spumarius (Hemiptera: Cercopidae) and its related species. Mol Phylogenet Evol 56:462–467
Shoemaker DD, Katju V, Jaenike J (1999) Wolbachia and the evolution of reproductive isolation between Drosophila recens and Drosophila subquinaria. Evolution 53:1157–1164
Shoemaker DD, Keller G, Ross KG (2003) Effects of Wolbachia on mtDNA variation in the two native fire ants Solenopsis invicta and S. richteri. Mol Ecol 12:1757–1771
Stewart JB, Beckenbach AT (2005) Insect mitochondrial genomics: the complete mitochondrial genome sequence of the meadow spittlebug Philaneus spumarius (Hemiptera: Auchenorrhyncha: Cercopoidae). Genome 48:46–54
Stouthamer R, Luck RF, Hamilton WD (1990) Antibiotics cause parthenogenetic Trichogramma to revert to sex. Proc Natl Acad Sci U S A 87:2424–2427
Stouthamer R, Breeuwer JAJ, Luck RF, Werren JH (1993) Molecular identification of microorganisms associated with parthenogenesis. Nature (London) 361:66–68
Stouthamer R, Breeuwer JAJ, Hurst GDD (1999) Wolbachia pipientis: microbial manipulator of reproduction. Ann Rev Microbiol 53:71–102
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood evolutionary distance and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Tishechkin DY (2013) Two new species of the genus Phileanus (Homoptera: Aphrophoridae) from Iran. Entomol Rev 93:73–76
Trpis M, Perrone JB, Reissig M, Parker KL (1981) Control of cytoplasmic incompatibility in the Aedes scuttelaris complex. J Hered 72:313–317
Turelli M (2010) Cytoplasmic incompatibility in populations with overlapping generations. Evolution 64:232–241
Van Borm S, Wenseleers T, Billen J, Boomsma JJ (2003) Cloning and sequencing of wsp encoding gene fragments reveals a diversity of co-infecting Wolbachia strains in Acromyrmex leafcutter ants. Mol Phylogenet Evol 26:102–109
Wade MJ, Stevens L (1985) Microorganism mediated reproductive isolation in flour beetles (genus Tribolium). Science 227:527–528
Wade MJ (2007) The co-evolutionary genetics of ecological communities. Nat Rev Genet 8:185–195
Watanabe M, Tagami Y, Miura K, Kageyama D, Stouthamer R (2012) Distribution patterns of Wolbachia endosymbionts in the closely related flower bugs of the genus Orius: implications for coevolution and horizontal transfer. Microb Ecol 64:537–545
Werren JH, Zhang W, Guo LR (1995) Evolution and phylogeny of Wolbachia: reproductive parasite of arthropod. Proc R Soc Lond B 161:55–63
Werren JH (1997) Biology of Wolbachia. Ann Rev Entomol 42:587–609
Werren JH, Windsor DW (2000) Wolbachia infection frequencies in insects: evidence of a global equilibrium. Proc R Soc Lond B 267:1277–1285
Werren JH, Baldo L, Clark ME (2008) Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6:741–751
West SA, Cook JM, Werren JH, Godfray HC (1998) Wolbachia in two insect host-parasitoid communities. Mol Ecol 7:1457–1465
Zabal-Aguirre M, Arroyo F, Bella JL (2010) Distribution of Wolbachia infection in Chorthippus parallelus populations within and beyond a Pyrenean hybrid zone. Heredity 104:174–184
Zabal-Aguirre M, Arroyo F, García-Hurtado J, de la Torre J, Hewitt GM, Bella JL (2014) Wolbachia effects in natural populations of Chorthippus parallelus from the Pyrenean hybrid zone. J Evol Biol 27:1136–1148
Zabalou S, Charlat S, Nirgianaki A, Lachaise D, Mercot H, Bourtzis K (2004) Natural Wolbachia infections in the Drosophila yakuba species complex do not induce cytoplasmic incompatibility but fully rescue the wRi modification. Genetics 167:827–834
Acknowledgments
We would like to thank two anonymous reviewers for their constructive remarks, which helped improve this article. Authors declare no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1a
a - e. Neighbor-joining phylogenic trees of Wolbachia strains in P. spumarius obtained with the use of MEGA5 for each MLST gene separately (GIF 49 kb)
Fig. S1b
(GIF 47 kb)
Fig. S1c
(GIF 47 kb)
Fig. S1d
(GIF 50 kb)
Fig. S1e
(GIF 46 kb)
Fig. S2a
a - e. Median-Joining networks of Wolbachia strains in P. spumarius obtained by SplitTree4 for each MLST gene separately (GIF 122 kb)
Fig. S2b
(GIF 185 kb)
Fig. S2c
(GIF 82 kb)
Fig. S2d
(GIF 108 kb)
Fig. S2e
(GIF 88 kb)
Table S1
MLST allelic profiles of Philaenus spumarius (DOC 148 kb)
Table S2
Hosts harboring similar strains of Wolbachia as P. spumarius. GB GenBank, SC sequence coverage, MI maximum identity (DOC 52 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lis, A., Maryańska-Nadachowska, A. & Kajtoch, Ł. Relations of Wolbachia Infection with Phylogeography of Philaenus spumarius (Hemiptera: Aphrophoridae) Populations Within and Beyond the Carpathian Contact Zone. Microb Ecol 70, 509–521 (2015). https://doi.org/10.1007/s00248-015-0570-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-015-0570-2