Abstract
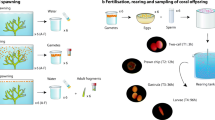
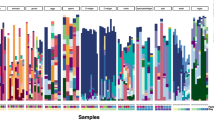
The oviparous sponge Ectyoplasia ferox is commonly found in Florida and the Bahamas. Every year in August and/or September about 6 days after a full moon, E. ferox will shed embryo-containing spawning material into the seawater from which hundreds to thousands of larvae will hatch per host individual. In order to investigate vertical microbial transmission in E. ferox, 16S rRNA gene library construction and denaturing gradient gel electrophoresis was employed. Microbial symbionts from six phyla and the unknown lineage SAUL were shown to be vertically transmitted. The identification of 21 VT clusters, of which 19 were situated within sponge-specific or sponge-coral-specific clusters, indicated that a large fraction of the symbiotic microbial consortium was present in the sexual reproductive stages. Spawning led to a 50 % reduction of microbial numbers in the adult sponge mesohyl. We furthermore provide the first evidence that the symbiotic microbial consortia of E. ferox were generally metabolically active within the reproductive stages. Finally, we propose E. ferox as a model system for vertical transmission owing to the ease of experimental access to all sexual reproductive stages, and to experimental tractability in the laboratory including the possibility of rearing symbiont-free juvenile sponges.









Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2006) At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71:7724–7736
Brusca RC, Brusca GJ (1990) Phylum Porifera: the sponges. In: Sinauer AD (ed) Invertebrates. Sinauer, Sunderland, pp 181–210
Cytryn E, van Rijn J, Schramm A, Gieseke A, de Beer D, Dror M (2005) Identification of bacteria potentially responsible for oxic and anoxic sulfide oxidation in biofilters of a recirculating mariculture system. Appl Environ Microbiol 71(10):6134–6141
DeCaralt S, Uriz MJ, Wijffels RH (2007) Vertical transmission and successive location of symbiotic bacteria during embryo development and larva formation in Corticium candelabrum (Porifera: Demospongiae). J Mar Biol Ass UK 87:1693–1699
Enticknap JJ, Kelly M, Peraud O, Hill RT (2006) Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl Environ Microbiol 72:3724–3732
Ereskovsky AV (2010) The comparative embryology of sponges. Springer, Heidelberg
Ereskovsky AV, Boury-Esnault N (2002) Cleavage pattern in Oscarella species (Porifera, Demospongiae, Homoscleromorpha), transmission of maternal cells and symbiotic bacteria. J Nat Hist 36:1761–1775
Ereskovsky AV, Gonobobleva E, Vishnyakov A (2005) Morphological evidence for vertical transmission of symbiotic bacteria in the viviparous sponge Halisarca dujardini Johnston (Porifera, Demospongiae, Halisarca). Mar Biol 146:869–875
Ereskovsky AV, Tokina D (2004) Morphology and fine structure of the swimming larvae of Ircinia oros (Porifera, Demospongiae, Dictyoceratida). Invertebr Reprod Dev 45:137–150
Ereskovsky AV, Willenz P (2008) Larval development in Guancha arnesenae (Porifera, Calcispongiae, Calcinea). Zoomorphology 127:175–187
Erwin PM, López-Legentil S, González-Pech R, Turon X (2012) A specific mix of generalists: bacterial symbionts in Mediterranean Ircinia spp. FEMS Microbiol Ecol 79(3):619–637
Fieseler L, Horn M, Wagner M, Hentschel U (2004) Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl Environ Microbiol 70:3724–3732
Friedrich AB, Hacker J, Fischer I, Proksch P, Hentschel U (2001) Temporal variations of the microbial community associated with the Mediterranean sponge Aplysina aerophoba. FEMS Microbiol Ecol 38:105–113
Giles E, Kamke J, Moitinho-Silva L, Taylor M, Hentschel U, Ravasi T, Schmitt S Bacterial community profiles in low microbial abundance sponges. FEMS Microbiol Ecol. doi:10.1111/j.1574-6941.2012.01467.x
Hentschel U, Fieseler L, Wehrl M, Gernert C, Steinert M, Hacker J, Horn M (2003) Microbial diversity of marine sponges. In: Müller WEG (ed) Molecular marine biology of sponges. Springer, Heidelberg, pp 60–88
Hentschel U, Hopke J, Horn M, Friedrich AB, Wagner M, Hacker J, Moore BS (2002) Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol 68:4431–4440
Hentschel U, Piel J, Degnan SM, Taylor MW (2012) Genomic insights into the marine sponge microbiome. Nat Rev Microbiol 10(9):641–654
Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC (2007) How rhizobial symbionts invade plants: the Sinorhizobium–Medicago model. Nat Rev Microbiol 5(8):619–633
Kamke J, Taylor MW, Schmitt S (2010) Activity profiles for marine sponge-associated bacteria obtained by 16S rRNA vs 16S rRNA gene comparisons. ISME 4(4):498–508
Kaye H (1991) Sexual reproduction in four Caribbean commercial sponges. II. Oogenesis and transfer of bacterial symbionts. Invertebr Reprod Dev 19:13–24
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, London, pp 115–175
Lee OO, Chui PY, Wong YH, Pawlik JR, Qian PY (2009) Evidence for vertical transmission of bacterial symbionts from adult to embryo in the Caribbean sponge Svenzea zeai. Appl Environ Microbiol 75(19):6147–6156
Levi C, Levi P (1976) Embryogenese de Chondrosia reniformis (Nardo), demosponge ovipare, et transmission des bacteries symbiotiques. Ann Sci Nat Zool 18:367–380
Leys SP, Ereskovsky AV (2006) Embryogenesis and larval differentiation in sponges. Canad J Zool 84:262–287
Lindquist N, Bolser R, Laing K (1997) Timing of larval release by two Caribbean demosponges. Mar Ecol Prog Ser 155:309–313
Ludwig W et al (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Maldonado M (2007) Intergenerational transmission of symbiotic bacteria in oviparous and viviparous demosponges, with emphasis on intracytoplasmically-compartmented bacterial types. J Mar Biol Ass UK 87:1701–1713
Maldonado M (2009) Embryonic development of verongid demosponges supports the independent acquisition of spongin skeletons as an alternative to the siliceous skeleton of sponges. Biol J Linnean Soc 97:427–447
Maldonado M, Riesgo A (2009) Gametogenesis, embryogenesis, and larval features of the oviparous sponge Petrosia ficiformis (Haplosclerida, Demospongiae). Mar Biol 156:2181–2197
Montalvo NF, Hill RT (2011) Sponge-associated bacteria are strictly maintained in two closely related but geographically distant sponge hosts. Appl Environ Microbiol 77(20):7207–7216
Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C (1998) Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans ADL, van Elsas JD, de Bruijn FJ (eds) Molecular microbial ecology, manual 3.4.4. Kluwer, Dordrecht
Nyholm SV, Stewart JJ, Ruby EG, McFall-Ngai MJ (2009) Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol 11(2):483–493
Oren M, Steindler L, Ilan M (2005) Transmission, plasticity and the molecular identification of cyanobacterial symbionts in the Red Sea sponge Diacarnus erythraenus. Mar Biol 148:35–41
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Gloeckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Rottmann M, Schroeder HC, Gramzow M, Renneisen K, Kurelec B, Dorn A, Friese U, Mueller WE (1987) Specific phosphorylation of proteins in pore complex-laminae from the sponge Geodia cydonium by the homologous aggregation factor and phorbol ester. Role of protein kinase C in the phosphorylation of DNA topoisomerase II. EMBO J 6:3939–3944
Sambrook J, Russell D (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schmitt S, Angermeier H, Schiller R, Lindquist N, Hentschel U (2008) Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl Environ Microbiol 74(24):7694–7708
Schmitt S, Hentschel U, Taylor MW (2012) Deep sequencing reveals diversity and community structure of complex microbiota in five Mediterranean sponges. Hydrobiologia 687:341–351
Schmitt S, Tsai P, Bell J, Fromont F, Ilan M, Lindquist NL, Perez T, Rodrigo A, Schupp PJ, Vacelet J, Webster N, Hentschel U, Taylor MW (2012) Assessing the complex sponge microbiota—core, variable, and species-specific bacterial communities in marine sponges. ISME J 6(3):564–576
Schmitt S, Wehrl M, Lindquist N, Weisz JB, Hentschel U (2008b) Morphological and molecular analyses of microorganisms in Caribbean reef adult sponges and in corresponding reproductive material. In: Custódio MR, Lôbo-Hajdu G, Hajdu E, Muricy G (eds) Porifera research biodiversity, innovation and sustainability. Série Livros 28. Museu Nacional, Rio de Janeiro, pp 561–568
Schmitt S, Weisz JB, Lindquist N, Hentschel U (2007) Vertical transmission of a phylogenetically complex microbial consortium in the viviparous sponge Ircinia felix. Appl Environ Microbiol 73(7):2067–2078
Sciscioli M, Lepore E, Gherardi M, Scalera Liaci L (1994) Transfer of symbiotic bacteria in the mature oocyte of Geodia sydonium (Porifera, Demospongiae): an ultrastructural study. Cah Biol Mar 35:471–478
Sharp KH, Eam B, Faulkner DJ, Haygood MG (2007) Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl Environ Microbiol 73:622–629
Steger D, Ettinger-Epstein P, Whalan S, Hentschel U, de Nys R, Wagner M, Taylor MW (2008) Diversity and mode of transmission of ammonia-oxidizing archaea in marine sponges. Environ Microbiol 10(4):1087–1094
Steindler L, Huchon D, Avni A, Ilan M (2005) 16S rRNA phylogeny of sponge-associated cyanobacteria. Appl Environ Microbiol 71(7):4127–4131
Taylor MW, Radax R, Steger D, Wagner M (2007) Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71:295–347
Usher K, Ereskovsky AV (2005) Larval development, ultrastructure and metamorphosis in Chondrilla australiensis Carter, 1873 (Demospongiae, Chondrosida, Chondrillidae). Invert Reprod Dev 47:51–62
Usher KM, Kuo J, Fromont J, Sutton DC (2001) Vertical transmission of cyanobacterial symbionts in the marine sponge Chondrilla australiensis (Demospongiae). Hydrobiologia 461:15–23
Vacelet J (1975) Étude en microscopie électronique de l’association entre bactéries et spongiaires du genre Verongia (Dictyoceratida). J Microsc Biol Cell 23:271–288
Webster NS, Taylor MW (2012) Marine sponges and their microbial symbionts: love and other relationships. Environ Microbiol 14:335–346
Webster NS, Taylor MW, Behnam F, Lücker S, Rattei T, Whalan S, Horn M, Wagner M (2010) Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ Microbiol 12(8):2070–2082
Wehrl M, Steinert M, Hentschel U (2007) Bacterial uptake by the marine sponge Aplysina aerophoba. Microb Ecol 53:355–365
Acknowledgments
We gratefully acknowledge the marine operations personnel of the National Undersea Research Center (NURC) of UNC Wilmington (Key Largo, FL), Hilde Angermeier (University of Würzburg) for excellent support during field work, and Alexander Ereskovsky (CNRS, Marseille, France) for valuable help in interpretation of the microscopic images. Research was supported by German Research Foundation (Deutsche Forschungsgmeinschaft) grant HE3299/1–3 to UH.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gloeckner, V., Lindquist, N., Schmitt, S. et al. Ectyoplasia ferox, an Experimentally Tractable Model for Vertical Microbial Transmission in Marine Sponges. Microb Ecol 65, 462–474 (2013). https://doi.org/10.1007/s00248-012-0142-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-012-0142-7