Abstract
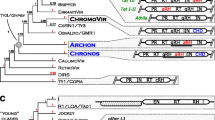
Retroviruses differ from retrotransposons due to their infective capacity, which depends critically on the encoded envelope. Some plant retroelements contain domains reminiscent of the env of animal retroviruses but the number of such elements described to date is restricted to angiosperms. We show here the first evidence of the presence of putative env-like gene sequences in a gymnosperm species, Pinus pinaster (maritime pine). Using a degenerate primer approach for conserved domains of RNaseH gene, three clones from putative envelope-like retrotransposons (PpRT2, PpRT3, and PpRT4) were identified. The env-like sequences of P. pinaster clones are predicted to encode proteins with transmembrane domains. These sequences showed identity scores of up to 30% with env-like sequences belonging to different organisms. A phylogenetic analysis based on protein alignment of deduced aminoacid sequences revealed that these clones clustered with env-containing plant retrotransposons, as well as with retrotransposons from invertebrate organisms. The differences found among the sequences of maritime pine clones isolated here suggest the existence of different putative classes of env-like retroelements. The identification for the first time of env-like genes in a gymnosperm species may support the ancestrality of retroviruses among plants shedding light on their role in plant evolution.





Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Berenyi M, Gichuki ST, Schmidt J, Burg K (2002) Ty1-copia retrotransposon-based S-SAP (sequence-specific amplified polymorphism) for genetic analysis of sweetpotato. Theor Appl Genet 105:862–869
Boeke JD, Corces VG (1989) Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol 43:403–434
Boeke JD, Stoye JP (1997) Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin JM, Hughes SH, Varmus HE (eds) retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 343–436
Chavanne F, Zhang D-X, Liaud M-F, Cerff R (1998) Structure and evolution of Cyclops: a novel giant retrotransposon of the Ty3/gypsy family highly amplified in pea and other legume species. Plant Mol Biol 37:363–375
Desset S, Conte C, Dimitri P, Calco V, Dastugue B, Vaury C (1999) Mobilization of two retroelements, ZAM and Idefix, in a novel unstable line of Drosophila melanogaster. Mol Biol Evol 16:54–66
Emori Y, Shiba T, Kanaya S, Inouye S, Yuki S, Saigo K (1985) The nucleotide sequences of copia and copia-related RNA in Drosophila virus-like particles. Nature 315:773–776
Favell AJ, Dunbar E, Anderson R, Pearce SR, Hartley R (1992) Ty–1 copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res 20:3639–3644
Friesen N, Brandes A, Heslop-Harrison J (2001) Diversity, origin and distribution of retrotransposons in conifers. Mol Biol Evol 18:1176–1188
Gallo SA, Finnegan CM, Viard M, Raviv Y, Dimitrov A, Rawat SS, Puri A, Durell S, Blumenthal R (2003) The HIV env-mediated fusion reaction. Biochim Biophys Acta Biomembr 1614:36–50
Gonzales P, Lessios HA (1999) Evolution of sea urchin retroviral-like (SURL) elements: evidence from 40 echinoid species. Mol Biol Evol 16:938–952
Hofmann K, Stoffel W (1993) TMbase—a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler 374:166
Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:W585–W587
Hull R (2001) Classifying reverse transcribing elements: a proposal and a challenge to the ICTV. Arch Virol 146:2255–2261
Jordan IK, Matyunina LV, McDonald JF (1999) Evidence for the recent horizontal transfer of long terminal repeat retrotransposon. Proc Natl Acad Sci USA 96:12621–12625
Kim FJ, Battini J-L, Manel N, Sitbon M (2004) Emergence of vertebrate retroviruses and envelope capture. Virology 318:183–191
Konieczny A, Voytas DF, Cummingst MP, Ausubel FM (1991) A Superfamily of Arabidopsis thaliana Retrotransposons. Genetics 127:801–809
Kumar A (1998) The evolution of plant retroviruses: moving to green pastures. Trends Plant Sci 3:371–374
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Laten HM, Majumdar A, Gaucher EA (1998) Sire-1, a copia Ty1-like retroelement from soybean, encodes a retroviral envelope-like protein. Proc Natl Acad Sci USA 95:6897–6902
Laten HM, Havecker ER, Farmer LM, Voytas DF (2003) Sire1, an endogenous retrovirus family from Glycine max, is highly homogeneous and evolutionarily young. Mol Biol Evol 20:1222–1230
Lerat E, Capy P (1999) Retrotransposons and retroviruses: analysis of the envelope gene. Mol Biol Evol 16:1198–1207
Malik HS, Eickbush TH (2001) Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res 11:1187–1197
Marco A, Marín I (2005) Retrovirus-like elements in plants. Recent Res Dev Plant Sci 3:1–10
Nakai K, Kanehisa M (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14:897–911
Neumann P, Pozárková D, Koblízková A, Macas J (2005) PIGY, a new plant envelope-class LTR retrotransposon. Mol Gen Genom 275:43–53
Pearce SR (2006) SIRE-1, a putative plant retrovirus is closely related to a legume Ty1-copia retrotransposon family. Cell Mol Biol Lett 12:120–126
Pearce SR, Stuart-Rogers C, Knox MR, Kumar A, Ellis THN, Flavell AJ (1999) Rapid isolation of plant Ty1-copia group retrotransposon LTR sequences for molecular marker studies. Plant J 19:711–717
Pélissier T, Tutois S, Deragon J-M, Tourmente S, Genestier S, Picard G (1995) Athila, a new retroelement from Arabidopsis thaliana. Plant Mol Biol 29:441–452
Peterson-Burch BD, Wright DA, Laten HM, Voytas DF (2000) Retroviruses in plants? Trends Genet 16:151–152
Rabson AB, Graves BJ (1997) Synthesis and processing of viral RNA. In: Coffin JM, Hughes SH, Varmus HE (eds) Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 205–261
Rost B, Casadio R, Fariselli P, Sander C (1995) Transmembrane helices predicted at 95% accuracy. Prot Sci 4:521–533
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schulman AH, Flavell AJ, Ellis THN (2004) The application of LTR retrotransposons as genetic markers in plants. In: Capy W (ed) Mobile genetic elements: protocols and genomic applications. Humana Press, Totowa, NJ, pp 145–173
Stuart-Rogers C, Flavell AJ (2001) The evolution of Ty1-copia group retrotransposons in gymnosperms. Mol Biol Evol 18:155–163
Suoniemi A, Tanskanen J, Schlman AH (1998) Gypsy-like retrotransposon are widespread in the plant kingdom. Plant J 13:699–705
Swanstrom R, Wills JW (1997) Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE (eds) Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 263–334
Syed NH, Sureshsundar S, Wilkinson MJ, Bhau BS, Cavalcanti JJV, Flavell AJ (2005) Ty1-copia retrotransposon-based SSAP marker development in cashew (Anacardium occidentale L.). Theor Appl Genet 110:1195–1202
Tanda S, Mullor JL, Corces VG (1994) The Drosophila tom retrotransposon encodes an envelope protein. Mol Cell Biol 14:5392–5401
van-Regenmortel MHV, Fauquet CM, Bishop DHL, Carsten EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR (2000) Virus taxonomy: seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA
Vicient CM, Kalendar R, Schulman AH (2001) Envelope-class retrovirus-like elements are widespread, transcribed and spliced, and insertionally polymorphic in plants. Genome Res 11:2041–2049
Vogt VM (1997) Retroviral virions and genomes. In: Coffin JM, Hughes SH, Varmus HE (eds) Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 27–69
Vos P, Hogers R, Bleeker M, Reijans M, Tvd Lee, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wright DA, Voytas DF (1998) Potential retroviruses in plants: Tat1 is related to a group of Arabidopsis thaliana Ty3/gypsy retrotransposons that encode envelope-like proteins. Genetics 149:703–715
Wright DA, Voytas DF (2001) Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res 12:122–131
Xiong Y, Eickbush TH (1990) Origin and evolution of retroelements based on their reverse transcriptase sequences. EMBO J 9:3353–3362
Yano ST, Panbehi B, Das A, Laten HM (2005) Diaspora, a large family of Ty3-gypsy retrotransposons in Glycine max, is an envelope-less member of an endogenous plant retrovirus lineage. BMC Evol Biol 5(30):1–14
Acknowledgments
The authors are grateful to Prof. Wanda Viegas and Prof. Leonor Morais-Cecílio for helpful discussions and critical readings of the manuscript. This research was supported by Fundação para a Ciência e Tecnologia (FCT), the III Framework Program of the EC, and POCI 2010 through grants SFRH/BPD/17902/2004 and SFRH/BD/32037/2006 and through project POCTI/AGR/46283/2002.
Author information
Authors and Affiliations
Corresponding author
Additional information
Célia Miguel and Margarida Rocheta contributed equally to this article.
Rights and permissions
About this article
Cite this article
Miguel, C., Simões, M., Oliveira, M.M. et al. Envelope-Like Retrotransposons in the Plant Kingdom: Evidence of Their Presence in Gymnosperms (Pinus pinaster). J Mol Evol 67, 517–525 (2008). https://doi.org/10.1007/s00239-008-9168-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-008-9168-3