Abstract
Purpose
Our purpose was to investigate the feasibility of pharmacy-initiated pharmacogenetic (PGt) screening in primary care with respect to patient willingness to participate, quality of DNA collection with saliva kits, genotyping, and dispensing data retrieved from the pharmacy.
Methods
Polypharmacy patients aged >60 years who used at least one drug with Anatomical Therapeutic Chemical (ATC) code N06AA01–N06AX19 (antidepressants), A02BC01–A02BC05 (proton-pump inhibitors), N05AA01–N05AH04 (antipsychotics), or C07AB02 (metoprolol) in the preceding 2 years were randomly selected. DNA was collected with saliva kits and genotyped for CYP2D6 and CYP2C19 with the AmpliChip. Pharmacy dispensing records were retrieved and screened for drugs interacting with the patient’s CYP2D6 and CYP2C19 genotype by using the evidence-based PGt guidelines from the Dutch Pharmacogenetics Working Group.
Results
Out of the 93 invited patients, 54 (58.1%) provided informed consent. Nine saliva samples (16.7%) contained too little DNA. Call rates for CYP2D6 and CYP2C19 were 93.3% and 100%, respectively. Frequencies of genotype-predicted phenotype were 2.4%, 38.1%, 54.8%, and 4.8% for CYP2D6 poor metabolizers (PM), intermediate metabolizers (IM), extensive metabolizers (EM), and ultrarapid metabolizers (UM) respectively. For CYP2C19 genotype-predicted phenotype, frequencies were 2.2%, 15.6%, and 82.2% for PM, IM, and EM, respectively.
Conclusions
This study shows that pharmacy-initiated PGt screening is feasible for a primary care setting.
Similar content being viewed by others
Introduction
Pharmacogenetics (PGt) promises an exciting approach to a more individualized drug therapy, ultimately leading to a more efficient and safer application of drugs. To date, many efforts within the field of PGt have been aimed at improving drug therapy with high-risk medications, such as those used within the field of oncology. By contrast, multiple PGt interventions for drug therapies with lower risk might also prove beneficial [1]. Yet the clinical use of genotyping prior to drug prescribing and dispensing is not widely practiced. [2] PGt information is accumulating rapidly, and it was reported that based on the available PGt information, it is possible to generate advice for nearly 100 drugs for patients with a completely sequenced genome. [3] In addition, Philips et al. reported that 16 of 27 (59%) drugs most commonly associated with adverse drug reactions (ADRs) were metabolized by a polymorphic gene. [4] A subsequent study published by Grice et al. investigated the frequency of use of these 16 PGt ADR-associated drugs in a primary care setting. [5] It was reported that 28.6% of patients took more than one PGt ADR-associated drug, indicating a high potential for PGt to make drug therapy safer. A disadvantage of that study was that it did not include drugs for which PGt testing is recommended to enhance efficacy instead of to avoid ADRs. Van Puijenbroek et al. reported their study aimed at gaining insight into the feasibility of informing physicians reporting ADRs to The Netherlands Pharmacovigilance Centre about possible PGt involvement and genotyping their patients. The authors reported that 39.5% of the reporting health-care professionals actually initiated genotyping. [6] This illustrates that incorporating PGt information in drug prescribing could increase safety of drug therapy.
Two large (inter)national initiatives aimed at providing PGt-based guidelines and recommendations concerning drug prescribing have been published. [7, 8] The Clinical Pharmacogenetics Implementation Consortium (CPIC) published its first guideline [9], with the goal of providing peer-reviewed, evidence-based, freely accessible guidelines for gene/drug pairs. CPIC published a list of 29 gene/drug pairs that were ranked highest in a survey of the American Society for Clinical Pharmacology and Therapeutics members in 2010. [7] The Dutch Pharmacogenetics Working group published updated evidence-based guidelines with PGt recommendations concerning 53 drugs. [8] From that group’s list of highest-ranked priority gene/drug pairs from CPIC and the article by Grice et al. [5], it can be observed that many drugs used in primary care are to some extent influenced by PGt, again illustrating a potential role for PGt in primary care.
The aim of our study was to test the feasibility of pharmacy-initiated PGt screening in primary care with respect to patient willingness to participate, quality of DNA collection with saliva kits, genotyping, and quality of dispensing data retrieved from pharmacy records.
Methods
Study setting
In The Netherlands, the vast majority of the population obtains their medication from only one community pharmacy, enabling collection of complete medication histories. [10] The pharmacy keeps an electronic patient record that covers all dispensing data. Polypharmacy patients were recruited from a community pharmacy located in the city of Leiden, The Netherlands. Patients were selected from the pharmacy records if they used at least one drug that is metabolized by CYP2D6 or CYP2C19 and at least four additional drugs in the preceding 2 years. Drugs were selected on their Anatomical Therapeutic Chemical (ATC) classification code [http://www.whocc.no/atc_ddd_index/]. To identify eligible ATC codes, we used textbooks, an academic Web site [http://medicine.iupui.edu/flockhart/table.htm], and a review article by Kirchheiner et al. [11–13]. Codes eligible for inclusion comprised N06AA01–N06AX19 (antidepressants), A02BC01–A02BC05 (proton-pump inhibitors), N05AA01–N05AH04 (antipsychotics), and ATC-code C07AB02 (metoprolol). Patients had to be ≥60 years at the start of the study. This age was chosen for the practical reason that elderly patients are more frequently at home, thus simplifying saliva collection.
Sample collection
Samples of 2 ml saliva were collected during a 30-min house visit. Samples were collected in the Oragene DNA self-collection kit (DNA Genotek Inc Ottawa, Ontario, Canada). A sample of 125 patients was drawn from the selected patients by randomization, with the aim of inviting 100 patients by mail and to finally obtain DNA samples from 50 patients. General practitioners were informed of our study prior to contacting the patient and asked to exclude patients with terminal disease status. Patients selected for inclusion received a letter from the pharmacy explaining the study background and objectives. Approximately 1–2 weeks after the letter was sent, patients were contacted by phone by one of the pharmacists participating in the study (EV). During this phone call, patients were asked if they agreed to participate. If so, a 30-min house visit was planned to collect informed consent and a saliva specimen. If the first attempt to contact a patient by phone was unsuccessful, a maximum of three consecutive attempts was made. After that, the patient was considered not willing to participate. The study was approved by the ethics committee of the Leiden University Medical Center. Informed consent was obtained from all participants.
Genotyping
DNA was extracted from saliva using the Oragene DNA self-collection kit according to the manufacturer’s instruction at the Leiden University Medical Center (Leiden, The Netherlands). DNA concentrations were measured with nanodrop (Isogen, Maarssen, The Netherlands) and diluted with water to a concentration of 10 ng/μl. The DNA was tested for 29 known polymorphisms in the CYP2D6 gene, including gene duplication and deletion, as well as two major polymorphisms in the CYP2C19 gene. Genotyping was performed at the Department of Clinical Chemistry of the Erasmus MC (Rotterdam, The Netherlands) using the AmpliChip CYP450 test (Roche Molecular Systems, Alameda, CA, USA) according to the manufacturer’s instruction. A genotype-predicted-phenotype (phenotype) was assigned to each patient. [8] For CYP2D6, intermediate metabolizers (IMs) were defined as patients carrying two decreased-activity (*9, *10, *17, *29, *36, *41) alleles or carrying one active (*1, *2, *33, *35) and one inactive (*3–*8, *11–*16, *19–*21, *38, *40, *42) allele, or carrying one decreased-activity (*9, *10, *17, *29, *36, *41) allele and one inactive (*3–*8, *11–*16, *19–*21, *38, *40, *42) allele. Poor metabolizers (PMs) were defined as patients carrying two inactive (*3–*8, *11–*16, *19–*21, *38, *40, *42) alleles. Ultrarapid metabolizers (UMs) were defined as patients carrying a gene duplication in the absence of inactive (*3–*8, *11–*16, *19–*21, *38, *40, *42) or decreased-activity (*9, *10, *17, *29, *36, *41) alleles. All other patients were considered extensive metabolizers (EMs). For CYP2C19, IMs were defined as patients with one active (*1) and one inactive (*2, *3) allele. PMs were defined as patients carrying two inactive alleles.
Medication analyses
Data were extracted from the pharmacy dispensing records; drugs for topical application were excluded. The number of unique prescribed drugs per patient was calculated as the number of unique ATC codes prescribed to each patient in the studied period of 2 years. For each unique prescribed drug, we checked whether a recommendation was available in the guidelines of the Dutch Pharmacogenetics Working Group of the Royal Dutch Association for the Advancement of Pharmacy. [8] These evidence-based guidelines contain a comprehensive evaluation of PGt gene–drug interaction involving 53 drugs and 11 genes. To illustrate the potential impact of PGt on primary care, the percentage of prescribed drugs with any PGt recommendation was calculated for each patient and in detail for PGt recommendations regarding CYP2D6 and CYP2C19. To evaluate whether patients with a non-EM CYP2D6 or CYP2C19 status had been empirically switched to non-CYP2D6 / CYP2C19 substrates, the percentage of CYP2D6/CYP2C19 substrates with a PGt recommendation of the total number of prescribed drugs was compared between EMs and non-EMs.
Statistical analysis
Student's t test was used to evaluate differences in the percentage of prescribed CYP2D6 and CYP2C19 substrates, and the number of drugs with a recommendation in the guideline of the Dutch Pharmacogenetics Working Group between EMs and non-EMs. A p value < 0.05 was regarded as significant. Statistical analysis was conducted with the SPSS statistical package (version 17.0, SPSS, Chicago, IL, USA).
Results
Patient response
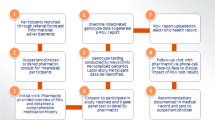
Five hundred and seven patients representing approximately 5% of the total registered patient population were prescribed at least one drug from the selected ATC codes and at least four additional drugs (Fig. 1). Of these patients, a random sample of 125 was selected: 22 patients were excluded because they visited a general practitioner that did not participate in the study; a further ten patients were excluded because of terminal disease status as judged by their general practitioner. Of the 93 invited patients, 54 (58.1%) provided informed consent. Twenty-two (23.7%) of the 93 invited patients refused to participate, and 17 (18.3%) could not be included for other reasons (Fig. 1). The mean age of included patients was 71 (range 60–91) years. Ethnicity was not routinely recorded, but all patients were of European ancestry as observed during saliva-sample collection (by EV). There were more women (61.1%) than men (38.9%) in the cohort. The percentage of invited men and women who agreed to participate was 54.1% and 65.6%, respectively (p = 0.29).
Genotyping
Nine saliva samples contained too little DNA for genotyping with the AmpliChip (< 50 ng). The call rate after a single run was 93.3% for CYP2D6 (three no-calls) and 100% for CYP2C19. As expected, the most frequent alleles were the CYP2D6*1 and CYP2D6*2, with a frequency of 0.35 and 0.21, respectively. The other functional allele, *35, had a prevalence of 0.10. The CYP2D6*4 allele was the most frequent zero-activity allele, followed by the *5 and *3 allele (0.17, 0.02, 0.01, respectively). The inactive alleles *41, *9, and *10 had a prevalence of 0.06, 0.04, and 0.02, respectively. Most patients (54.8%) were predicted to have the EM phenotype, followed by IM (38.1%), UM (4.8%), and PM (2.4%). Prevalence of the CYP2C19* 1 and *2 alleles was 0.90 and 0.10, respectively. No carriers of CYP2C19*3 allele were found. One patient was homozygous for the CYP2C19*2 allele and therefore categorized as PM. Seven patients were heterozygous carriers of the CYP2C19*2 allele and therefore categorized as IM. All other patients were considered EMs.
Medication history
The mean number of unique prescribed drugs per patient was 15.2 [95% confidence interval (CI) 13.4–17.1] in the 2-year study period, with an average of 4.6 (95% CI 4.0–5.2) prescriptions per unique prescribed drug. The percentage of CYP2D6 substrates with a PGt recommendation of the total number of prescribed drugs was not different between CYP2D6 non-EMs, and patients with a predicted CYP2D6 EM phenotype, with 4.70% and 4.86%, respectively (p = 0.95). For CYP2C19, similar results were found, with 8.53% and 9.26% of the prescribed drugs having a PGt recommendation for non-EMs and EMs, respectively (p = 0.74). For patients with a predicted CYP2D6 or CYP2C19, non-EM phenotype detailed information about prescriptions for drugs for which a CYP2D6 or CYP2C19 PGt recommendation was available are provided in Table 1. The CYP2D6 PM used no drugs with a CYP2D6 PGt recommendation. Of the CYP2D6 UMs, one patient used no drugs with a CYP2D6 PGt recommendation, and one patient used codeine 10 mg three times daily and paroxetine 20 mg once daily. The codeine was limited to a single prescription only. The latter patient was also predicted to be a CYP2C19 IM and used clopidogrel 75 mg once daily and omeprazole 40 mg twice daily. The reduced metabolic capacity for CYP2C19 and the drug–drug interaction between clopidogrel and omeprazole both lead to reduced formation of the active metabolite of clopidogrel and subsequent increased risk for therapeutic failure [14, 15]. The CYP2D6 IMs were often prescribed tramadol or codeine, drugs that both are expected to have a reduced analgesic effect in CYP2D6 IMs. Metoprolol was also often prescribed, but according to the guidelines of the Dutch Pharmacogenetics Working Group only requires a dose adjustment when used for heart failure. The CYP2C19 IMs and PM were mostly prescribed proton-pump inhibitors, which do not require a dose adjustment, as decreased metabolism results in increased therapeutic efficacy.
As we were interested in the potential impact of PGt on primary care, the medication history of all patients was further evaluated for drugs metabolized by enzymes other than CYP2D6 and CYP2C19. On average patients used 2.3 (15.9%, 95% CI 1.9–2.7) drugs for which a recommendation of the Dutch Pharmacogenetics Working Group was available. The most frequently prescribed drugs with a PGt recommendation were the proton-pump inhibitors, followed by phenprocoumon and metoprolol (Table 2). The number of drugs with a therapeutic recommendation did not differ between EM and non-EMs, with 2.2 versus 2.5 drugs for CYP2D6 EMs and non-EMs, respectively (p = 0.48), and 2.5 versus 1.8 drugs for CYP2C19 EMs and non-EMs ,respectively (p = 0.24).
Discussion
The available evidence of genetic variants with clinical relevance regarding both efficacy and toxicity of drug therapy is accumulating rapidly. Elaborating on this information, multiple initiatives to develop clear-cut therapeutic guidelines translating available evidence to therapeutic recommendations have been initiated [7, 8, 16]. However, there is little information regarding the potential impact of these recommendations in primary care. This study indicates that a majority of patients is willing to participate in a PGt screening study and that pharmacy-initiated PGt screening is feasible in a primary care setting. Of the invited patients, 58.1% was willing to participate in our study. This is a relatively large percentage given the fact that screening was not directly related to a clinical problem. For patients presenting with an ADR or lack of therapeutic effect, willingness to participate in PGt screening may be even higher. Age has also been reported to be of influence on the attitude toward PGt testing, with younger patients being more likely to be optimistic about the usefulness of PGt testing. [17] Therefore, willingness to participate may be higher in a population younger than 60 years. On the other hand, we collected DNA samples during a visit at the patients’ homes. Collection by mail may result in lower response rates.
More women than men participated in our study, although there was no statistically significant difference in the willingness to participate between the sexes. Sex differences concerning PGt testing have been reported, with female patients being more likely to have concerns regarding the possible negative consequences of PGt testing and being less willing to participate than male patients. [17, 18] However, the finding that more women participated in this study might simply be explained by the fact that from 65 years onward, the female to male ratio is starting to increase due to higher life expectancy of women.
The reported allele, genotype, and phenotype frequencies are comparable with previously reported results obtained with the AmpliChip [19] or other methods, such as polymerase chain reaction restriction fragment length polymorphism (PCR- RFLP) in comparable populations of mainly white individuals. [20, 21]. These results indicate that no selection bias occurred and that our patient cohort is representative.
Our study is limited in that we used a dichotomous outcome parameter to categorize drugs as either being with or without a PGt recommendation in the Dutch Pharmacogenetics Working Group guidelines. All drugs for which, according to the guidelines, a gene–drug interaction is present and an action is required were put in a single category. This is an oversimplification because the guidelines provide many different types of recommendations, e.g., advice to adjust the dose, be extra alert to diminished therapeutic efficacy, or increased risk for an ADR. Also, the recommendation depends on the metabolism of the drug, e.g., does the drug have active metabolites or is it a prodrug? For example, codeine is used for both pain and cough. The analgesic effect requires the formation of morphine by CYP2D6. Therefore, CYP2D6 UMs will require a dose reduction for an equal pain reduction compared with EMs, whereas no dose reduction is required for the effect on cough. Dose adjustments were not investigated in this feasibility study. In addition, patients who used at least one drug metabolized by CYP2D6 or CYP2C19 were selected for inclusion. As a result, this study does not provide quantitative estimates of the incidence of the use of drugs with a PGt recommendation for CYP2D6 or CYP2C19 in the Dutch PGt guidelines. The study was not designed to—and therefore does not—provide direct evidence that the use of PGt recommendations results in improved efficacy or decreased toxicity. That requires further study.
We identified some potential pitfalls for PGt screening in primary care as performed in this study. First, for a number of patients, production of the required 2 ml saliva was difficult. As the included patients used at least five different medications each, this might be explained by the use of anticholinergic medication or other drugs that cause a dry mouth. Indeed, five of the nine patients who failed to provide sufficient DNA used this type of medication. Secondly, the no-call rate for the AmpliChip was 6.7% for CYP2D6. This is relatively high when compared with results reported by other groups. In a study with 158 breast cancer patients treated with tamoxifen, Borges et al. reported a no-call rate of 0.7%. Serrano et al. reported a no-call rate of 1.6% in 182 Italian breast cancer patients treated with tamoxifen. Both studies used DNA extracted from whole blood for genotyping. However, in a large study of 4,532 psychiatric patients, the no-call rate for CYP2D6 with the AmpliChip was 6.0% after three rounds of testing. [22] In that study, it was first attempted to genotype patients with DNA extracted from buccal swabs or saliva specimens. When saliva/buccal DNA failed to provide a genotype, blood DNA was tested. The no-call rate after primary genotyping was as high as 13%. According to the authors, this was mainly the result of DNA collection with buccal swabs, as this DNA tends to be contaminated by bacterial DNA. [22] DNA extracted from saliva has been reported to be of high quality and a suitable alternative to blood DNA. [23] In our laboratory, we compared genotyping results of an additional set of 24 DNA samples for which both blood and saliva were available. The no-call rate for saliva was 4% higher compared with the no-call rate for blood samples, with 3/24 and 2/24 for saliva vs. blood, respectively. Although these results are inconclusive and further investigation is required, they may indicate that DNA extracted from saliva results in slightly less successful genotyping with the AmpliChip.
According to the guidelines of the Dutch Pharmacogenetics Working Group, approximately 5–10% of drugs prescribed to patients with aberrant CYP2D6 or CYP2C19 metabolism require action, such as dose adjustment or extra awareness for an ADR. [8] In our study, there was no difference in the percentage of prescribed CYP2D6 and CYP2C19 substrates between EMs and non-EMs of CYP2D6 or CYP2C19. This suggests that physicians have not empirically identified patients with aberrant metabolism, e.g., by switching patients with side effects to medications that were not substrate for CYP2D6 or CYP2C19. From Table 1 it can also be observed that patients with a non-EM phenotype were prescribed regular drug dosages. Available data do not allow an in-depth analysis of switching behavior, as this requires a complete medication history, including the fist prescription, and not just a cross-sectional period of 2 years.
Of the total number of prescribed medications, 15.9% had a PGt therapeutic recommendation in the Dutch guidelines. This number is comparable with a previously published estimate that 15–20% of prescribed drugs are metabolized by genetically polymorphic enzymes [24]. However, our results should be interpreted with extreme caution, as one of the inclusion criteria was the use of at least one drug metabolized by CYP2D6 or CYP2C19. Therefore, the number of drugs with a PGt recommendation may be overestimated in our data.
In conclusion, this study shows that pharmacy-initiated PGt screening is feasible for a primary care setting.
References
Altman RB (2011) Pharmacogenomics: "noninferiority" is sufficient for initial implementation. Clin Pharmacol Ther 89:348–350
Wang L, McLeod HL, Weinshilboum RM (2011) Genomics and drug response. N Engl J Med 364:1144–1153
Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, Pushkarev D, Neff NF, Hudgins L, Gong L, Hodges LM, Berlin DS, Thorn CF, Sangkuhl K, Hebert JM, Woon M, Sagreiya H, Whaley R, Knowles JW, Chou MF, Thakuria JV, Rosenbaum AM, Zaranek AW, Church GM, Greely HT, Quake SR, Altman RB (2010) Clinical assessment incorporating a personal genome. Lancet 375:1525–1535
Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W (2001) Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA 286:2270–2279
Grice GR, Seaton TL, Woodland AM, McLeod HL (2006) Defining the opportunity for pharmacogenetic intervention in primary care. Pharmacogenomics 7:61–65
Van Puijenbroek PE, Conemans J, van Grootheest K (2009) Spontaneous ADR reports as a trigger for pharmacogenetic research: a prospective observational study in the Netherlands. Drug Saf 32:255–264
Relling MV, Klein TE (2011) CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther 89:464–467
Swen JJ, Nijenhuis M, de Boer A, Grandia L, Maitland-van der Zee AH, Mulder H, Rongen GA, van Schaik RH, Schalekamp T, Touw DJ, van der Weide J, Wilffert B, Deneer VH, Guchelaar HJ (2011) Pharmacogenetics: from bench to byte- an update of guidelines. Clin Pharmacol Ther 89:662–673
Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui CH, Yee SW, Stein CM, Carrillo M, Evans WE, Klein TE (2011) Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther 89:387–391
Buurma H, Bouvy ML, De Smet PA, Floor-Schreudering A, Leufkens HG, Egberts AC (2008) Prevalence and determinants of pharmacy shopping behaviour. J Clin Pharm Ther 33:17–23
Hansten P, Horn J (2004) The top 100 drug interactions. A guide to patient management H&H Publications
Cavallari L, Ellingrod V, Kolesar J (2006) Lexi-Comp's Pharmacogenomics Handbook, 2nd edition Edition, Lexi Comp
Kirchheiner J, Nickchen K, Bauer M, Wong ML, Licinio J, Roots I, Brockmoller J (2004) Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry 9:442–473
Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias W, Braunwald E, Sabatine MS (2009) Cytochrome p-450 polymorphisms and response to clopidogrel. NEnglJMed 360:354–362
Ho PM, Maddox TM, Wang L, Fihn SD, Jesse RL, Peterson ED, Rumsfeld JS (2009) Risk of adverse outcomes associated with concomitant use of clopidogrel and proton pump inhibitors following acute coronary syndrome. JAMA 301:937–944
Swen JJ, Wilting I, de Goede AL, Grandia L, Mulder H, Touw DJ, de Boer A, Conemans JM, Egberts TC, Klungel OH, Koopmans R, van der Weide J, Wilffert B, Guchelaar HJ, Deneer VH (2008) Pharmacogenetics: from bench to byte. Clin Pharmacol Ther 83:781–787
Rogausch A, Prause D, Schallenberg A, Brockmoller J, Himmel W (2006) Patients' and physicians' perspectives on pharmacogenetic testing. Pharmacogenomics 7:49–59
van Wieren-de Wijer DB, Maitland-van der Zee AH, de Boer A, Kroon AA, de Leeuw PW, Schiffers P, Janssen RG, Psaty BM, van Duijn CM, Stricker BH, Klungel OH (2009) Reasons for non-response in observational pharmacogenetic research. PharmacoepidemiolDrug Saf 18:665–671
Borges S, Desta Z, Li L, Skaar TC, Ward BA, Nguyen A, Jin Y, Storniolo AM, Nikoloff DM, Wu L, Hillman G, Hayes DF, Stearns V, Flockhart DA (2006) Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther 80:61–74
Tamminga WJ, Wemer J, Oosterhuis B, de Zeeuw RA, de Leij LF, Jonkman JH (2001) The prevalence of CYP2D6 and CYP2C19 genotypes in a population of healthy Dutch volunteers. Eur J Clin Pharmacol 57:717–722
Sachse C, Brockmoller J, Bauer S, Roots I (1997) Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. Am J Hum Genet 60:284–295
de Leon J, Susce MT, Johnson M, Hardin M, Maw L, Shao A, Allen AC, Chiafari FA, Hillman G, Nikoloff DM (2009) DNA microarray technology in the clinical environment: the AmpliChip CYP450 test for CYP2D6 and CYP2C19 genotyping. CNSSpectr 14:19–34
Rylander-Rudqvist T, Hakansson N, Tybring G, Wolk A (2006) Quality and quantity of saliva DNA obtained from the self-administrated oragene method–a pilot study on the cohort of Swedish men. Cancer Epidemiol Biomarkers Prev 15:1742–1745
Evans WE, Relling MV (1999) Pharmacogenomics: translating functional genomics into rational therapeutics. Science 286:487–491
Acknowledgments
No additional funding was received.
Conflict of interest
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Swen, J.J., van der Straaten, T., Wessels, J.A.M. et al. Feasibility of pharmacy-initiated pharmacogenetic screening for CYP2D6 and CYP2C19 . Eur J Clin Pharmacol 68, 363–370 (2012). https://doi.org/10.1007/s00228-011-1130-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00228-011-1130-4