Abstract
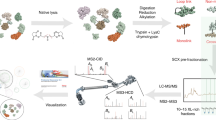
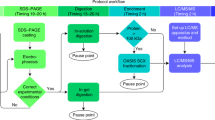
Chemical cross-linking/mass spectrometry (MS) is gradually developing into a routine method to investigate protein conformation and to decipher protein interaction networks. To increase identification rates of the frequently low abundant cross-linked products in LC/MS/MS experiments, fast and reliable sample preparation protocols are indispensable. We present simplified solid phase extraction methods using C18/SCX StageTips and mixed-mode OASIS MCX cartridges for a single-step enrichment of cross-linked products prior to LC/MS/MS analysis. Our improved protocols result in 3.5 to 4.6 times higher numbers of cross-link identifications for the model protein bovine serum albumin compared to non-processed samples.



Similar content being viewed by others
Abbreviations
- BS3 :
-
Bis(sulfosuccinimidyl)suberate
- BSA:
-
Bovine serum albumin
- CID:
-
Collision-induced dissociation
- ESI:
-
Electrospray ionization
- HEPES:
-
4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid
- HPLC:
-
High-performance liquid chromatography
- LC:
-
Liquid chromatography
- LTQ:
-
Linear ion trap (Thermo Fisher Scientific)
- MCX:
-
Mixed mode cation exchange (Waters)
- MeOH:
-
Methanol
- MS:
-
Mass spectrometry
- MS/MS:
-
Tandem mass spectrometry
- RP:
-
Reversed phase
- SCX:
-
Strong cation exchange chromatography
- SEC:
-
Size exclusion chromatography
- SPE:
-
Solid phase extraction
- TFA:
-
Trifluoroacetic acid
References
Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein–protein interactions. Mass Spectrom Rev. 2006;25:663–82.
Rappsilber J. The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J Struct Biol. 2011;173:530–40.
Holding AN. XL-MS: protein cross-linking coupled with mass spectrometry. Methods. 2015;89:54–63.
Arlt C, Ihling CH, Sinz A. Structure of full-length p53 tumor suppressor probed by chemical cross-linking and mass spectrometry. Proteomics. 2015;15:2746–55.
Sinz A, Arlt C, Chorev D, Sharon M. Chemical cross-linking and native mass spectrometry: a fruitful combination for structural biology. Protein Sci. 2015;24:1193–209.
Yang B, Wu Y-J, Zhu M, Fan S-B, Lin J, Zhang K, et al. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9:904–6.
Hoopmann MR, Zelter A, Johnson RS, Riffle M, MacCoss MJ, Davis TN, et al. Kojak: efficient analysis of chemically cross-linked protein complexes. J Proteome Res. 2015;14:2190–8.
Liu F, Rijkers DTS, Post H, Heck AJR. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat Methods. 2015;12:1179–84.
Götze M, Pettelkau J, Schaks S, Bosse K, Ihling CH, Krauth F, et al. StavroX—a software for analyzing crosslinked products in protein interaction studies. J Am Soc Mass Spectrom. 2012;23:76–87.
Götze M, Pettelkau J, Fritzsche R, Ihling CH, Schäfer M, Sinz A. Automated assignment of MS/MS cleavable cross-links in protein 3D-structure analysis. J Am Soc Mass Spectrom. 2015;26:83–97.1.
Kalkhof S, Ihling C, Mechtler K, Sinz A. Chemical cross-linking and high-performance Fourier transform ion cyclotron resonance mass spectrometry for protein interaction analysis: application to a calmodulin/target peptide complex. Anal Chem. 2005;77:495–503.
Pearson KM, Pannell LK, Fales HM. Intramolecular cross-linking experiments on cytochrome C and ribonuclease A using an isotope multiplet method. Rapid Commun Mass Spectrom. 2002;16:149–59.
Müller DR, Schindler P, Towbin H, Wirth U, Voshol H, Hoving S, et al. Isotope-tagged cross-linking reagents. A new tool in mass spectrometric protein interaction analysis. Anal Chem. 2001;73:1927–34.
Arlt C, Götze M, Ihling CH, Hage C, Schäfer M, Sinz A. Integrated workflow for structural proteomics studies based on cross-linking/mass spectrometry with an MS/MS cleavable cross-linker. Anal Chem. 2016;88:7930–7.
Vellucci D, Kao A, Kaake RM, Rychnovsky SD, Huang L. Selective enrichment and identification of azide-tagged cross-linked peptides using chemical ligation and mass spectrometry. J Am Soc Mass Spectrom. 2010;21:1432–45.
Tan D, Li Q, Zhang M-J, Liu C, Ma C, Zhang P, Ding Y-H, Fan S-B, Tao L, Yang B, Li X, Ma S, Liu J, Feng B, Liu X, Wang H-W, He S-M, Gao N, Ye K, Dong M-Q, Lei X. Trifunctional cross-linker for mapping protein-protein interaction networks and comparing protein conformational states. Elife. 2016;5.
Chu F, Mahrus S, Craik CS, Burlingame AL. Isotope-coded and affinity-tagged cross-linking (ICATXL): an efficient strategy to probe protein interaction surfaces. J Am Chem Soc. 2006;128:10362–3.
Kang S, Mou L, Lanman J, Velu S, Brouillette WJ, Prevelige PE. Synthesis of biotin-tagged chemical cross-linkers and their applications for mass spectrometry. Rapid Commun Mass Spectrom. 2009;23:1719–26.
Back JW, de Jong L, Muijsers AO, de Koster CG. Chemical cross-linking and mass spectrometry for protein structural modeling. J Mol Biol. 2003;331:303–13.
Fritzsche R, Ihling CH, Götze M, Sinz A. Optimizing the enrichment of cross-linked products for mass spectrometric protein analysis. Rapid Commun Mass Spectrom. 2012;26:653–8.
Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, et al. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 2010;29:717–26.
Rinner O, Seebacher J, Walzthoeni T, Mueller LN, Beck M, Schmidt A, et al. Identification of cross-linked peptides from large sequence databases. Nat Methods. 2008;5:315–8.
Leitner A, Reischl R, Walzthoeni T, Herzog F, Bohn S, Förster F, Aebersold R. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol Cell Proteomics. 2012;11:M111.014126.
Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–906.
Acknowledgements
AS is supported by the DFG (project Si 867/15-2) and the region of Saxony Anhalt. The authors are indebted to Mr. Thomas Piotrowski for constructing the SPE apparatus (ESM, Fig. S12).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1206 kb)
Rights and permissions
About this article
Cite this article
Schmidt, R., Sinz, A. Improved single-step enrichment methods of cross-linked products for protein structure analysis and protein interaction mapping. Anal Bioanal Chem 409, 2393–2400 (2017). https://doi.org/10.1007/s00216-017-0185-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-017-0185-1