Abstract
The process of protein digestion is a critical step for successful protein identification in proteomic analysis. Many efforts have been dedicated to enhancing the digestion efficiency for sufficient digestion. Among these approaches, protein complete denaturation with denaturants is a common process for better digestion. However, the removal of denaturants was tedious or would cause protein loss and other problems. In this work, a feasible digestion approach, immobilized protein digestion (IPD), based on covalent binding has been developed. Proteins can be completely denatured and immobilized on the surface of functional materials by covalent binding to form a monolayer. Subsequently, varieties of denaturants or contaminants would be removed thoroughly by washing. To achieve fast immobilization and high digestion efficiency, different functional materials and denaturants were selected. Compared with traditional in-solution digestion, the method achieved a prominent increase in identified peptides numbers and sequence coverage of proteins. Data analysis also showed that covalent binding could evidently decrease enzymatic missed cleavage for various protein sequences. Furthermore, possible peptide losses due to covalent binding were also investigated. Also, it has been proved to be efficient for complex biological sample digestion.

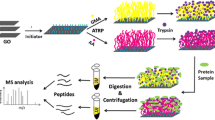
Workflow of the IPD method, including protein denaturation, immobilization, digestion, and identification






Similar content being viewed by others
References
Fields S. Proteomics-proteomics in genomeland. Science. 2001;291(5507):1221–4.
Zhang YY, Fonslow BR, Shan B, Baek MC, Yates JR. Protein analysis by shotgun/bottom-up proteomics. Chem Rev. 2013;113:2343–94.
Switzar L, Giera M, Niessen WMN. Protein digestion: an overview of the available techniques and recent developments. J Proteome Res. 2013;12:1067–77.
Zougman A, Selby PJ, Banks RE. Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. Proteomics. 2014;14:1006–10.
Ma J, Zhang LH, Liang Z, Shan YC, Zhang YK. Immobilized enzyme reactors in proteomics. Trends Anal Chem. 2011; 30 (5).
Liu JY, Lin S, Qi DW, Deng CH, Yang PY, Zhang XM. On-chip enzymatic microreactor using trypsin-immobilized superparamagnetic nanoparticles for highly efficient proteolysis. J Chromatogr A. 2007;1176(1–2):169–77.
Li Y, Zhang XM, Deng CH. Functionalized magnetic nanoparticles for sample preparation in proteomics and peptidomics analysis. Chem Soc Rev. 2013;42:8517.
Lin S, Yao GP, Qi DW. Fast and efficient proteolysis by microwave-assisted protein digestion using trypsin-immobilized magnetic silica microspheres. Anal Chem. 2008;80(10):3655–65.
Yao GP, Deng CH, Zhang XM, Yang PY. Efficient tryptic proteolysis accelerated by laser radiation for peptide mapping in proteome analysis. Angew Chem Int Ed. 2010;49:8185–9.
Beyazit S, Ambrosini S, Marchyk N, Palo E, Kale V, Soukka T. Versatile synthetic strategy for coating upconverting nanoparticles with polymer shells through localized photopolymerization by using the particles as internal light sources. Angew Chem Int Ed. 2014;53:8919–23.
Changa CF, Truongb QD, Chena JR. Graphene as excellent support for rapid and efficient near infrared-assisted tryptic proteolysis. Colloids Surf B: Biointerfaces. 2013;104:221–8.
Ge H, Bao H, Zhang L, Chen G. Immobilization of trypsin on miniature incandescent bulbs for infrared-assisted proteolysis. Anal Chim Acta. 2014;845(3):77–84.
Xu Y, Mehl JT, Bakhtiar R, Woolf EJ. Immunoaffinity purification using anti-PEG antibody followed by two-dimensional liquid chromatography/tandem mass spectrometry for the quantification of a PEGylated therapeutic peptide in human plasma. Anal Chem. 2010;82:6877–86.
Callipo L, Caruso G, Foglia P, Gubbiotti R, Samperi R, Laganà A. Immunoprecipitation on magnetic beads and liquid chromatography–tandem mass spectrometry for carbonic anhydrase II quantification in human serum. Anal Biochem. 2010;400:195–202.
Wisniewski JR, Mann M. Consecutive proteolytic digestion in an enzyme reactor increases depth of proteomic and phosphoproteomic analysis. Anal Chem. 2012;84:2631–7.
Dator RP, Gaston KW, Limbach PA. Multiple enzymatic digestions and Ion mobility separation improve quantification of bacterial ribosomal proteins by data independent acquisition liquid chromatography–mass spectrometry. Anal Chem. 2014;86:4264–70.
Vandermarliere E, Mueller M, Martens L. Getting intimate with trypsin, the leading protease in proteomics. Mass Spectrom Rev. 2013;32:453–65.
Setou M, Hayasaka T, Shimma S, Sugiura Y, Matsumoto M. Protein denaturation improves enzymatic digestion efficiency for direct tissue analysis using mass spectrometry. Appl Surf Sci. 2008;255:1555–9.
Wang HB, Hu GF, Zhang YQ, Yuan Z, Zhao X, Zhu Y. Optimization and quality assessment of the post-digestion 18O labeling based on urea for protein denaturation by HPLC/ESI-TOF mass spectrometry. J Chromatogr B. 2010;878(22):1946–52.
Crowell AMJ, Stewart EJ, Take ZS, Doucette AA. Critical assessment of the spectroscopic activity assay for monitoring trypsin activity in organic-aqueous solvent. Anal Biochem. 2013;435:131–6.
Santos HM, Mota C, Lodeiro C, Moura I, Isaac I, Capelo JL. An improved clean sonoreactor-based method for protein identification by mass spectrometry-based techniques. Talanta. 2008;77:870–5.
Hervey WJ, Strader MB, Hurst GB. Comparison of digestion protocols for microgram quantities of enriched protein samples. J Proteome Res. 2007;6:3054–61.
Zhang N, Chen R, Young N, Wishart D. Comparison of SDS- and methanol-assisted protein solubilization and digestion methods for Escherichia coli membrane proteome analysis by 2-D LC-MS/MS. Proteomics. 2007;7:484–93.
Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62.
Acknowledgments
This work was supported by the National Research Projects (2016YFA0501402, 2012YQ12004409, and 2013CB911201).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The use of the mouse liver samples for research was approved by the Ethics Committee of Zhongshan Hospital and the Biomedical Sciences, Fudan University. All authors declare that they are in compliance with the ethical standards.
Authors’ contributions
All authors have given approval to the final version of the manuscript.
Rights and permissions
About this article
Cite this article
Qi, Q., Yan, G., Deng, C. et al. A novel protocol for enzymatic digestion based on covalent binding by protein immobilization. Anal Bioanal Chem 408, 8437–8445 (2016). https://doi.org/10.1007/s00216-016-9964-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9964-3