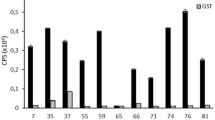
Abstract
Fast and reliable diagnostic assays are required for a resilient detection of clinical infections or biothreat-relevant pathogens. While PCR has proven to be the gold standard for nucleic acid detection, the identification of pathogen particles is still challenging and depends on the availability of well-characterized, chemically stable, and selective recognition molecules. Here, we report the screening of a phage display random peptide library for vaccinia virus-binding peptides. The identified peptide was extensively characterized using peptide-probe ELISA, surface plasmon resonance, nLC-MS/MS, Western Blot, peptide-based immunofluorescence assay, and electron microscopy. Following identification, the phage-free, synthetic peptide, designated αVACVpep05, was shown to bind to vaccinia virus and other orthopoxviruses. We can demonstrate that the highly conserved orthopoxvirus surface protein D8 is the interaction partner of αVACVpep05, thus enabling the peptide to bind to other orthopoxviruses, including cowpox virus and monkeypox virus, viruses that cause clinically relevant zoonotic infections in humans. The process of phage display-mediated peptide identification has been optimized intensively, and we provide recommendations for the identification of peptides suitable for the detection of further pathogens. The peptide described here was critically characterized and seems to be a promising reagent for the development of diagnostic platforms for orthopoxviruses. We believe that our results will help to promote the development of alternative, nonantibody-based synthetic detection molecules for further pathogens.






Similar content being viewed by others
References
Lim DV, Simpson JM, Kearns EA, Kramer MF (2005) Current and developing technologies for monitoring agents of bioterrorism and biowarfare. Clin Microbiol Rev 18(4):583–607. doi:10.1128/CMR.18.4.583-607.2005
Kodadek T, Reddy MM, Olivos HJ, Bachhawat-Sikder K, Alluri PG (2004) Synthetic molecules as antibody replacements. Acc Chem Res 37(9):711–718. doi:10.1021/ar030145l
Ngundi MM, Kulagina NV, Anderson GP, Taitt CR (2006) Nonantibody-based recognition: alternative molecules for detection of pathogens. Expert Rev Proteomics 3(5):511–524. doi:10.1586/14789450.3.5.511
Petrenko V (2003) Phage display for detection of biological threat agents. J Microbiol Methods 53(2):253–262. doi:10.1016/s0167-7012(03)00029-0
Willats WGT (2002) Phage display: practicalities and prospects. Plant Mol Biol 50:837–854
Szardenings M (2003) Phage display of random peptide libraries: applications, limits, and potential. J Recept Signal Transduct Res 23(4):307–349. doi:10.1081/RRS-120026973
Wang X, Li G, Ren Y, Ren X (2011) Phages bearing affinity peptides to bovine rotavirus differentiate the virus from other viruses. PLoS ONE 6(12):e28667. doi:10.1371/journal.pone.0028667
Wang C, Sun X, Suo S, Ren Y, Li X, Herrler G, Thiel V, Ren X (2013) Phages bearing affinity peptides to severe acute respiratory syndromes-associated coronavirus differentiate this virus from other viruses. J Clin Virol. doi:10.1016/j.jcv.2013.04.002
Rogers JD, Ajami NJ, Fryszczyn BG, Estes MK, Atmar RL, Palzkill T (2013) Identification and characterization of a Peptide affinity reagent for detection of noroviruses in clinical samples. J Clin Microbiol 51(6):1803–1808. doi:10.1128/JCM.00295-13
Lavilla M, De Luis R, Perez MD, Calvo M, Sanchez L (2009) Selection of high affine peptide ligands for detection of Clostridium Tyrobutyricum spores. J Microbiol Methods 79(2):214–219. doi:10.1016/j.mimet.2009.09.008
Rao SS, Mohan KV, Gao Y, Atreya CD (2013) Identification and evaluation of a novel peptide binding to the cell surface of Staphylococcus aureus. J Microbiol Res 168(2):106–112. doi:10.1016/j.micres.2012.07.004
Morton J, Karoonuthaisiri N, Stewart LD, Oplatowska M, Elliott CT, Grant IR (2013) Production and evaluation of the utility of novel phage display-derived peptide ligands to Salmonella spp. for magnetic separation. J Appl Microbiol 115(1):271–281. doi:10.1111/jam.12207
Alibek K (2004) Smallpox: a disease and a weapon. Int J Infect Dis 8(Suppl 2):S3–8. doi:10.1016/j.ijid.2004.09.004
Essbauer S, Pfeffer M, Meyer H (2010) Zoonotic poxviruses. Vet Microbio 140(3–4):229–236. doi:10.1016/j.vetmic.2009.08.026
Sonntag M, Muhldorfer K, Speck S, Wibbelt G, Kurth A (2009) New adenovirus in bats, Germany. Emerg Infect Dis 15(12):2052–2055. doi:10.3201/eid1512.090646
Kurth A, Wibbelt G, Gerber HP, Petschaelis A, Pauli G, Nitsche A (2008) Rat-to-elephant-to-human transmission of cowpox virus. Emerg Infect Dis 14(4):670–671. doi:10.3201/eid1404.070817
Isaacs SN (2004) Vaccinia virus and poxvirology : methods and protocols. Methods Mol Biol, vol 269. Humana, Totowa, NJ
Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2007) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1(6):2856–2860
Rich RL, Quinn JG, Morton T, Stepp JD, Myszka DG (2010) Biosensor-based fragment screening using FastStep injections. Anal Biochem 407(2):270–277. doi:10.1016/j.ab.2010.08.024
Laue M (2010) Electron microscopy of viruses. Methods Cell Biol 96:1–20. doi:10.1016/S0091-679X(10)96001-9
Chang TH, Chang SJ, Hsieh FL, Ko TP, Lin CT, Ho MR, Wang I, Hsu ST, Guo RT, Chang W, Wang AH (2013) Crystal structure of vaccinia viral A27 protein reveals a novel structure critical for its function and complex formation with A26 protein. PLoS Pathog 9(8):e1003563. doi:10.1371/journal.ppat.1003563
Su HP, Singh K, Gittis AG, Garboczi DN (2010) The structure of the poxvirus A33 protein reveals a dimer of unique C-type lectin-like domains. J Virol 84(5):2502–2510. doi:10.1128/JVI.02247-09
Perdiguero B, Blasco R (2006) Interaction between vaccinia virus extracellular virus envelope A33 and B5 glycoproteins. J Virol 80(17):8763–8777. doi:10.1128/JVI.00598-06
Matho MH, Maybeno M, Benhnia MR, Becker D, Meng X, Xiang Y, Crotty S, Peters B, Zajonc DM (2012) Structural and biochemical characterization of the vaccinia virus envelope protein D8 and its recognition by the antibody LA5. J Virol 86(15):8050–8058. doi:10.1128/JVI.00836-12
Brown E, Senkevich TG, Moss B (2006) Vaccinia virus F9 virion membrane protein is required for entry but not virus assembly, in contrast to the related L1 protein. J Virol 80(19):9455–9464. doi:10.1128/JVI.01149-06
Van Vliet K, Mohamed MR, Zhang L, Villa NY, Werden SJ, Liu J, McFadden G (2009) Poxvirus proteomics and virus-host protein interactions. Microbio Mol Biol Rev : MMBR 73(4):730–749. doi:10.1128/MMBR.00026-09
Smith GP, Petrenko VA (1997) Phage display. Chem Rev 97(2):391–410
Hsiao JC, Chung CS, Chang W (1999) Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J Virol 73(10):8750–8761
Choi SK (2004) Synthetic multivalent molecules: concepts and biomedical applications. Wiley, Hoboken, NJ
Marik J, Lam KS (2005) Peptide and small-molecule microarrays. Methods Mol Biol 310:217–226
Acknowledgments
The authors are grateful to P. Wojciech Dabrowski for programming the “Library Insert Finder” software and to Andreas Kurth for providing us with adenovirus. We also thank Ursula Erikli for copy-editing. This work was funded within the BMBF/VDI-financed BiGRUDI network of the Robert Koch Institute (Berlin).
Conflict of interest
The authors declare no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Lilija Miller and Janine Michel contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1080 kb)
Rights and permissions
About this article
Cite this article
Miller, L., Michel, J., Vogt, G. et al. Identification and characterization of a phage display-derived peptide for orthopoxvirus detection. Anal Bioanal Chem 406, 7611–7621 (2014). https://doi.org/10.1007/s00216-014-8150-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-8150-8