Abstract
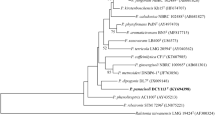
Strain DCY85T and DCY85-1T, isolated from rhizosphere of ginseng, were rod-shaped, Gram-reaction-negative, strictly aerobic, catalase positive and oxidase negative. 16S rRNA gene sequence analysis revealed that strain DCY85T as well as DCY85-1T belonged to the genus Burkholderia and were closely related to Burkholderia fungorum KACC 12023T (98.1 and 98.0 % similarity, respectively). The major polar lipids of strain DCY85T and DCY85-1T were phosphatidylethanolamine, one unidentified aminolipid and two unidentified phospholipids. The major fatty acids of both strains are C16:0, C18:1 ω7c and summed feature 3 (C16:1 ω6c and/or C16:1 ω7c). The predominant isoprenoid quinone of each strain DCY85T and DCY85-1T was ubiquinone (Q-8) and the G+C content of their genomic DNA was 66.0 and 59.4 mol%, respectively, which fulfill the characteristic range of the genus Burkholderia. The polyamine content of both DCY85T and DCY85-1T was putrescine. Although both DCY85T and DCY85-1T have highly similar 16S rRNA and identical RecA and gyrB sequences, they show differences in phenotypic and chemotaxonomic characteristics. DNA–DNA hybridization results proved the consideration of both strains as two different species. Based on the results from our polyphasic characterization, strain DCY85T and DCY85-1T are considered novel Burkholderia species for which the name Burkholderia ginsengiterrae sp. nov and Burkholderia panaciterrae sp. nov are, respectively, proposed. An emended description of those strains is also proposed. DCY85T and DCY85-1T showed antagonistic activity against the common root rot pathogen of ginseng, Cylindrocarpon destructans. The proposed type strains are DCY85T (KCTC 42054T = JCM 19888T) and DCY85-1T (KCTC 42055T = JCM 19889T).

Similar content being viewed by others
References
Anzai Y, Kudo Y, Oyaizu H (1997) The phylogeny of the genera Chryseomonas, Flavimonas, and Pseudomonas supports synonymy of these three genera. Int J Syst Bacteriol 47:249–251
Busse HJ, Auling G (1988) Polyamine pattern as a chemotaxonomic marker within the Proteobacteria. Syst Appl Microbiol 11:1–8
Coenye T, Laevens S, Willems A, Ohlen M, Hannant W, Govan JRW, Gillis M, Falsen E, Vandamme P (2001) Burkholderia fungorum sp. nov. and Burkholderia caledonica sp. nov., two new species isolated from the environment, animals and human clinical samples. Int J Syst Evol Microbiol 51:1099–1107
Coenye T, Henry D, Speert DP, Vandamme P (2004) Burkholderia phenoliruptrix sp. nov., to accommodate the 2,4, 5-trichlorophenoxyacetic acid and halophenol-degrading strain AC1100. Syst Appl Microbiol 27:623–627
Collins MD (1985) Isoprenoid quinone analyses in bacterial classification and identification. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics (Society for Applied Bacteriology Technical Series no. 20). Academic Press, London, pp 267–287
Cowan ST, Steel KJ (1974) Manual for the identification of medical bacteria. Cambridge University Press, Cambridge
Da K, Nowa KJ, Flinn B (2011) Potato cytosine methylation and gene expression changes induced by a beneficial bacterial endophyte, Burkholderia phytofirmans strain PsJN. Plant Phys Biochem J 50:24–34
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Gillis M, Van TV, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez MP (1995) Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol 45:274–289
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hankins A (2009) Producing and Marketing Wild Simulated Ginseng in Forest and Agroforestry Systems. Produced by Communications and Marketing, College of Agriculture and Life Sciences, Virginia Polytechnic Institute and State University
Howieson JG, Yates RJ, Foster K, Real D, Besier B (2008) Prospects for the future use of legumes. In: Dilworth MJ, James EK, Sprent JI, Newton WE (eds) Nitrogen-fixing leguminous symbioses. Elsevier, London, UK, pp 363–394
Kim HB, Park MJ, Yang HC, An DS, Jin HZ, Yang DC (2006) Burkholderia ginsengisoli sp. nov., a β-glucosidase-producing bacterium isolated from soil of a ginseng field. Int J Syst Bacteriol 56:2529–2533
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Lane DJ (1991) 16S/23S rRNA sequencing. In: Goodfellow M, Chichester, Stackebrandt E (eds) Nucleic acid techniques in bacterial systematics. Wiley, London, pp 115–176
Lee SS (2007) Korean ginseng (ginseng cultivation), Korean ginseng and T. Research institute, pp 18–40
Leung KW, Wong AS (2010) Pharmacology of ginsenosides: a literature review. Chin Med J. doi:10.1186/1749-8546-5-20
Lu P, Zheng LQ, Sun JJ, Liu HM, Li SP, Hong Q, Li WJ (2012) Burkholderia zhejiangensis sp. nov., a methyl-parathiondegrading bacterium isolated from a wastewater-treatment system. Int J Syst Evol Microbiol 62:1337–1341
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Mishra RP, Tisseyre P, Melkonian R, Chaintreuil C, Miche´ L, Klonowska A, Gonzalez S, Bena G, Laguerre G, Moulin L (2012) Genetic diversity of Mimosa pudicarhizobial symbionts in soils of French Guiana: investigating the origin and diversity of Burkholderia phymatum and other beta-rhizobia. FEMS Microbiol Ecol 79:487–503
Prescott LM, Harley JP (2001) The effects of chemical agents on bacteria II: antimicrobial agents (Kirby–Bauer method). In: Prescott LM, Harley JP (eds) Laboratory Exercises in Microbiology, 5th edn. McGraw-Hill Inc, New York, pp 57–262
Rahman M, Punja ZK (2005) Factors influencing development of root rot on ginseng caused by Cylindrocarpon destructans. Phytopathol J 95(12):1381–1390
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990). Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. MIDI Inc, Newark
Sheu SY, Chou JH, Bontemps C, Elliott GN, Gross E, Junior FBDR, Melkonian R, Moulin L, James EK, Sprent JI, Young JP, Chen WM (2013) Burkholderia diazotrophica sp. nov., isolated from root nodules of Mimosa spp. Int J Syst Evol Microbiol 63:435–441
Spilker T, Baldwin A, Bumford A, Dowson CG, Mahenthiralingam E, LiPuma JJ (2009) Expanded multilocus sequence typing for Burkholderia species. J Clin Microbiol 47:2607–2610
Suárez-Moreno ZR, Coutinho BG, Mendonça-Previato L, Previato L, James EK, Venturi V (2012) Common features of environmental and potentially beneficial plant-associated Burkholderia. Microbial Ecol 63:249–266
Taibi G, Schiavo MR, Gueli MC, Rindina PC, Muratore R, Nicotra CM (2000) Rapid and simultaneous high–performance liquid chromatography assay of polyamines and monoacetylpolyamines in biological specimens. J Chromatogr Biomed Sci Appl 745:431–437
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tian Y, Kong BH, Liu SL, Li CL, Yu R, Liu L, Li YH (2013) Burkholderia grimmiae sp. nov., isolated from a xerophilous moss (Grimmiamontana). Int J Syst Evol Microbiol 63:2108–2113
Vandamme P, Opelt K, Knochel N, Berg C, Schonmann S, Brandt E, Eberl L, Falsen E, Berg G (2007) Burkholderia bryophila sp. nov. and Burkholderia megapolitana sp. nov., moss-associated species with antifungal and plant-growth-promoting properties. Syst Evol Microbiol 57:2228–2235
Vandamme P, Brandt ED, Houf K, Salles JF, Elsas JDV, Spilker T, LiPuma JJ (2013) Burkholderia humi sp. nov., Burkholderia choica sp. nov., Burkholderia telluris sp. nov., Burkholderia terrestris sp. nov., and Burkholderia udeis sp. nov.: Burkholderia glathei-like bacteria from soil and rhizosphere soil. Syst Evol Microbiol. doi:10.1099/ijs.0.048900-0
Vanlaere E, van der Meer JR, Falsen E, Salles JF, Brandt ED, Vandamme P (2008) Burkholderia sartisoli sp. nov., isolated from a polycyclic aromatic hydrocarbon-contaminated soil. Int J Syst Evol Microbiol 58:420–423
Wayne L, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore EC, Murray RGE, Stackebrandt E, Starr MP, Truper HG (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M (1992) Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol 36:1251–1275
Zhao X, Gao J, Song C, Fang Q, Wang N, Zhao T, Liu D, Zhou Y (2012) Fungal sensitivity to and enzymatic deglycosylation of ginsenosides. Phytochem J 78:65–71
Acknowledgments
This research was supported by Korea Institute of Planning & Evaluation for Technology in Food, Agriculture, Forestry & Fisheries (KIPET NO: 313038-03-1-SB010) and Next-Generation BioGreen 21 Program (SSAC, Grant#: PJ009529032014), Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Erko Stackebrandt.
The GenBank/EMBL/DDBJ accession number for the 16S rRNA, gyrB and recA gene sequence of strain DCY85T and DCY85-1T are KF915802, KF999960, KM501455, KM501454, KM495734 and KM495735, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
203_2014_1075_MOESM1_ESM.tif
Supplementary Fig. S1. Neighbor-joining tree based on the recA gene sequences (approx. 566 bp) of DCY 85T and DCY 85-1T strains with other Burkholderia species. Bootstrap values after 1,000 replicates are expressed as percentages, values less than 50 % are not shown. Bar shows 0.1 substitutions per nucleotide position. (TIFF 619 kb)
203_2014_1075_MOESM2_ESM.tif
Supplementary Fig. S2. Neighbor-joining tree based on the gyrB gene sequences (approx. 690 bp) of DCY 85T and DCY 85-1T strains with reference species. Bootstrap values after 1,000 replicates are expressed as percentages, values less than 50 % are not shown. Bar shows 0.02 substitutions per nucleotide position. (TIFF 536 kb)
203_2014_1075_MOESM3_ESM.tif
Supplementary Fig. S3. Transmission electron microscope (TEM) pictures of both; a, DCY85T and b, DCY85-1T. (TIFF 1623 kb)
203_2014_1075_MOESM4_ESM.tif
Supplementary Fig. S4. Two-dimensional TLC of the total polar lipids of; a, B. ginsengiterrae DCY85T; b, B. panaciterrae DCY85-1T; c, B. fungorum KACC 12023T, stained with 5 % ethanolic molybdophosphoric acid. Abbreviations: PE, phosphatidylethanolamine; (AL), unidentified amino lipids; (PL), unidentified phospholipids and (L), unidentified polar lipids. (TIFF 1476 kb)
203_2014_1075_MOESM5_ESM.tif
Supplementary Fig. S5. Antagonistic activity of DCY85T and DCY85-1T along with B. fungorum KACC 12023T against; (a–d) F. solani; a, control; b, B. fungorum KACC 12023T; c, strain DCY 85T; d, strain DCY 85-1T. (e–h) F. oxysporum; e, control; f, B. fungorum KACC 12023T; g, DCY 85T; h, DCY 85-1T. (i–l) B. cinerea; i, control; j, B. fungorum KACC 12023T; k, DCY 85T; l, DCY 85-1T. (m–p) C. gloeosporioides; m, control; n, B. fungorum KACC 12023T; o, DCY 85T; p, DCY 85-1T. (q–t) C. destructans; q, control; r, B. fungorum KACC 12023T; s, DCY 85T and t, DCY 85-1T. The growth of the pathogens were recorded after 10 days except C. destructans, after 20 days. (TIFF 4892 kb)
Rights and permissions
About this article
Cite this article
Farh, M.EA., Kim, YJ., Van An, H. et al. Burkholderia ginsengiterrae sp. nov. and Burkholderia panaciterrae sp. nov., antagonistic bacteria against root rot pathogen Cylindrocarpon destructans, isolated from ginseng soil. Arch Microbiol 197, 439–447 (2015). https://doi.org/10.1007/s00203-014-1075-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-014-1075-y