Abstract
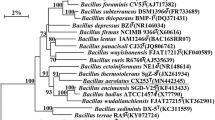
A novel thermophilic, Gram-staining positive bacterium, designated DX-2T, was isolated from the anode biofilm of a microbial fuel cell. Cells of the strain were oxidase positive, catalase positive, facultative anaerobic, motile rods. The isolate grew at 30–60 °C (optimum 50 °C) and pH 5–9 (optimum pH 8–8.5). The pairwise 16S rRNA gene sequence similarities showed that strain DX-2T was most closely related to Bacillus fumarioli LMG 17489T (96.2 %), B. firmus JCM 2512T (96.0 %) and B. foraminis DSM 19613T (95.7 %). Phylogenetic analysis based on 16S rRNA gene sequences showed that strain DX-2T formed a cluster with B. smithii (95.5 %) and B. infernus (94.9 %). The genomic G+C content of DX-2T was 43.7 mol%. The predominant respiratory quinone was MK-7. The polar lipids consisted of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine and unknown phospholipids. The major cellular fatty acid was iso-C16:0. Based on its phenotypic characteristics, chemotaxonomic features, and results of phylogenetic analysis, the strain was identified to represent a distinct novel species in the genus Bacillus, and the name proposed is B. thermophilum sp. nov. The type strain is DX-2T (=CCTCC AB2012194T = KCTC 33128T).

Similar content being viewed by others
References
Bae SS, Lee J-H, Kim S-J (2005) Bacillus alveayuensis sp. nov., a thermophilic bacterium isolated from deep-sea sediments of the Ayu Trough. Int J Syst Evol Microbiol 55:1211–1215
Baker GC, Smith JJ, Cowan DA (2003) Review and reanalysis of domain-specific 16S primers. J Microbiol Methods 55:541–555
Boon DR, Liu Y, Zhao ZJ, Balkwill DL, Drake GR, Stevens TO, Aldrich HC (1995) Bacillus infernus sp. nov., an Fe(III)- and Mn(IV)-reducing anaerobe from the deep terrestrial subsurface. Int J Syst Bacteriol 45:441–448
Breznak JA, Costilow RN (1994) Physicochemical factors in growth. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 137–154
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Dong X, Cai M (2001) Determination of biochemical properties. Manual for the systematic identification of general bacteria. Science Press, Beijing, pp 364–398
Fahmy F, Flossdorf J, Claus D (1985) The DNA base composition of the type strains of the genus Bacillus. Syst Appl Microbiol 6:60–65
Felsenstein J (1981) Evolutionary trees from DNA sequence: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Heyrman J, Rodríguez-Díaz M, Devos J, Felske A, Logan NA, Vos PD (2005) Bacillus arenosi sp. nov., Bacillus arvi sp. nov. and Bacillus humi sp. nov., isolated from soil. Int J Syst Evol Microbiol 55:111–117
Kämpfer P (1994) Limits and possibilities of total fatty acid analysis for classification and identification of Bacillis species. Syst Appl Microbiol 17:86–98
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeom YS, Lee JH, Yi H, Won S, Chun H (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Logan NA, Lebbe L, Hoste B, Goris J, Forsyth G, Heyndrickx M, Murray BL, Syme N, Wynn-Williams D, De Vos P (2000) Aerobic endospore-forming bacteria from geothermal environments in northern Victoria Land, Antarctica, and Candlemas Island, South Sandwich archipelago, with the proposal of Bacillus fumarioli sp. nov. Int J Syst Evol Microbiol 50:1741–1753
Logan NA, Berge O, Bishop AH, Busse HJ, De Vos P, Fritze D, Heyndrickx M, Kämpfer P, Rabinovitch L et al (2009) Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int J Syst Evol Microbiol 59:2114–2121
McCarthy AJ, Cross T (1984) A taxonomic study of Thermomonospora and other monosporic actinomycetes. J Gen Microbiol 130:5–25
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty-acid and polar lipid-composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Microbiol 47:87–95
Nakamura LK, Blumenstock I, Claus D (1988) Taxonomic study of Bacillus coagulans Hammer 1915 with proposal for Bacillus smithii sp.nov. Int J Syst Bacteriol 38:63–73
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. Newark, DE: MIDI
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tamaoka J, Katayama-Fujimura Y, Kuraishi H (1983) Analysis of bacterial menaquinone mixtures by high performance liquid chromatography. J Appl Bacteriol 54:31–36
Tamura K, Peterson D, Peterson N, Stecher G, Nei M et al (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tiago I, Pires C, Mendes V, Morais PV, da Costa MS, Veríssimo A (2006) Bacillus foraminis sp. nov., isolated from a nonsaline alkaline groundwater. Int J Syst Evol Microbiol 56:2571–2574
Vaishampayan P, Probst A, Krishnamurthi S, Ghosh S, Osman S, Mcdowall A, Ruckmani A, Mayilra S, Venkateswaran K (2010) Bacillus hornechiae sp. nov., isolated from a spacecraft-assembly clean room. Int J Syst Evol Microbiol 60:1031–1037
Acknowledgments
This work was supported by the Key Projects of the National Science & Technology Pillar Program of China (2012BAD14B16) and the Science and Technology Planning Project of Guangdong Province, China (2011B030900003, 2011A090200038 and 2011A030600003).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Erko Stackebrandt.
Jia Tang and Guiqin Yang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tang, J., Yang, G., Wen, J. et al. Bacillus thermophilum sp. nov., isolated from a microbial fuel cell. Arch Microbiol 196, 629–634 (2014). https://doi.org/10.1007/s00203-014-1001-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-014-1001-3