Abstract
The emerging discipline of plant phenomics aims to measure key plant characteristics, or traits, though as yet the set of plant traits that should be measured by automated systems is not well defined. Methods capable of recovering generic representations of the 3D structure of plant shoots from images would provide a key technology underpinning quantification of a wide range of current and future physiological and morphological traits. We present a fully automatic approach to image-based 3D plant reconstruction which represents plants as series of small planar sections that together model the complex architecture of leaf surfaces. The initial boundary of each leaf patch is refined using a level set method, optimising the model based on image information, curvature constraints and the position of neighbouring surfaces. The reconstruction process makes few assumptions about the nature of the plant material being reconstructed. As such it is applicable to a wide variety of plant species and topologies, and can be extended to canopy-scale imaging. We demonstrate the effectiveness of our approach on real images of wheat and rice plants, an artificial plant with challenging architecture, as well as a novel virtual dataset that allows us to compute distance measures of reconstruction accuracy. We also illustrate the method’s potential to support the identification of individual leaves, and so the phenotyping of plant shoots, using a spectral clustering approach.
Similar content being viewed by others
1 Introduction
In recent years, a growing recognition that the tools available to study the genetic structure of plants (the genotype) have outpaced those supporting analysis of plant structure and function (the phenotype) has lead to increased demand for new plant measurement methods. The emerging discipline of plant phenomics aims to extract quantitative measurements of key plant characteristics—traits—from image and sensor data. The resulting information is vital to efforts to understand plant growth and development and to ensure global food security in the face of climate change, resource depletion and an increasing population.
While a variety of approaches to plant shoot phenotyping have been proposed [1–5], there is as yet no clear definition as to the set of traits that should be sought. Where a set of traits are recovered [1], extending these traits to other plant species, and further to the general case of plant phenotyping, may prove challenging. Individual genetic variations might affect any aspect of the physical plant. Against this background, generic measurement and description methods are particularly valuable: the ability to construct rich descriptions of the 3D structure of plant shoots from images would underpin quantification of a wide range of current and future traits [6, 7].
Plants, however, provide a particularly challenging subject, with large amounts of self-occlusion, and, depending on plant species, leaves that lack the texture necessary to perform robust feature matching, either to separate leaves from one another, or locate specific leaves across multiple views. To overcome this, where image-based modelling approaches are successful, they have often involved user interaction [2].
Automatic methods can be classified as either top-down or bottom-up. Top-down approaches attempt to simplify the task by solving a model refinement problem. An existing model is adjusted to fit the image data, so that the new plant representation is consistent with what is observed. Quan et al. [2] and Ma et al. [8] take this approach, obtaining an ideal leaf model from a single leaf, and then fitting it to all other leaves in the scene. By adapting an existing model, topological inconsistency (such as the self-intersection of leaf surfaces) is avoided, but this comes at the expense of generality. Alenyà et al. [3] guides the segmentation of laser range data using planar or curved-quadratic surface models; however, this approach extends only to the refinement of point cloud data, without reconstructing leaf surfaces.
Bottom-up methods rely only on the observed pixel data. Silhouette-based methods [4, 9], and approaches derived from them [1], segment each image independently to identify the boundary of the object of interest. These regions are combined to determine the maximum possible object size consistent with the images presented to the algorithm, the photo hull [10]. Where the number of input images is high, the resulting model will be a good approximation to the true plant structure. However, as the scene becomes increasingly complex, for example with increasing numbers of leaves, larger plants, or multiple plants, the discrepancy between true object and model will increase.
Correspondence-based methods identify feature points independently in each of a set of images, then match those features between views. Knowledge of the cameras’ positions and orientations allow 3D locations of matched features to be computed. The method in [5] extracts the centre lines of wheat plants from two orthogonal viewpoints, improving reliability where single images would fail. This work does not, however, complete the 3D structure of each plant, preserving only the centre line of each leaf after skeletonisation.
Image-based modelling algorithms are widely applicable to a variety of subjects. Their generality can, however, become a limitation, where the representations they produce may be unsuitable for direct use in a given situation. The volumetric data structures produced by silhouette-based methods, for example, are static: the size and position of the voxels are defined early in the process and are difficult to change. While measurements of, e.g. height and volume are easily made from volumetric descriptions, estimating motion, e.g. of leaves moving in the breeze is extremely difficult. Similarly, point clouds can be used to calculate density and distributions of plant material, but cannot immediately be used, e.g. in leaf phenotyping applications, where a surface-based representation is required.
This paper describes a fully automatic, bottom-up approach to image-based 3D plant reconstruction that is applicable to a wide variety of plant species and topologies. The method is accurate, providing a true representation of the original plant, and produces data in a form that can support both trait measurement and modelling techniques such as forward ray tracing [11]. Our approach is outlined, and discussed in the context of photosynthesis modelling, in [12]. Here we present the technical details of the method and examine its ability to support plant phenotyping.
An initial 3D point cloud is first described by a set of planar patches, each representing a small section of plant material, usually a segment of leaf. Image noise and the complexity of the plant will, however, typically lead to missing areas of leaf material, and poorly defined surface boundaries. The initial surface estimate then is refined into a more accurate plant model, where the boundary of each surface patch is optimised based on the available image information, and positional information obtained from neighbouring surfaces.
The reconstruction process makes few assumptions about the nature of the plant material being reconstructed; by representing each leaf as a series of small planar sections, the complete leaf surface itself can take any reasonable shape. While our approach currently assumes plants are generally green, a modular design to the surface refinement function means any reasonable appearance model could be used in place of this. For example, an infra-red camera in a lab environment would produce a robust appearance model to use in place of RGB images. The generality of our technique allows it to be scaled to scenes involving multiple plants, and even plant canopies. However, the focus of this paper is on the accurate reconstruction of single plants of varying species.
2 Plant reconstruction
2.1 Input point cloud
The reconstruction algorithm described in this paper uses an initial point cloud estimate as a basis for the growth of plant surfaces in three dimensions. Numerous software- and hardware-based techniques exist to obtain point representations of objects. We have chosen to make use of a software-based technique, patch-based multi-view stereo (PMVS) [13]. This approach reconstructs dense point clouds from any calibrated image set, and is not restricted to plant data. However, by including robust visibility constraints, it is well suited to plant datasets that contain large amounts of occlusion. Let \(\{X_{i}\}^{n}_{i=1}\) be the set of all points in an input cloud of size n. We identify the co-ordinate system used by the point cloud, and the resulting reconstruction, as “world” co-ordinates. An individual point \(p \in X\) in world co-ordinates is represented as a 3D vector \(\varvec{w}\).
A requirement of both PMVS and our reconstruction approach is that the intrinsic and extrinsic camera parameters be known. We use the VisualSFM [14] system to perform automatic camera calibration. Any number of arbitrary camera positions may be calibrated using VisualSFM, and calibration is performed quickly. However, as it is based on SIFT features [15], the approach is not suitable for images with insufficient texture and feature information. This is particularly problematic within plant datasets, where leaves may have few suitable feature points. In our real plant datasets, the surrounding scene provides an adequate feature set for correspondence. In our artificial plant dataset, a highly textured calibration target is used, and in our virtual dataset camera parameters are extracted automatically without the need for calibration. We have found in our experiments that the calibration performed within VisualSFM is sufficiently accurate to drive PMVS, and our method. Where the intrinsic parameters of the camera are known, for example, where the model and lens are kept constant, it is possible to replace VisualSFM calibration with a more robust technique, which may improve accuracy.
We capture \(N_\mathrm{cam}\) images of the scene from \(N_\mathrm{cam}\) locations to obtain a set of images \(\{I_{i}\}^{N_\mathrm{cam}}_{i=1}\). Associated with each camera location is a perspective projection matrix, based on a standard pinhole camera model [16], derived from the calibration information output by VisualSFM. For a given world point, there is a perspective projection function, \(\mathcal {V}_{i}\), that maps onto a point in a specific camera co-ordinate frame, given by the 2D vector \(\varvec{v}\). This gives a set of functions \(\{\mathcal {V}_{j}(\varvec{w}):\mathbb {R}^{3}\rightarrow \mathbb {R}^{2}\}^{N_\mathrm{cam}}_{j=1}\), where j is the index of the input image and associated camera geometry. Once in camera co-ordinates, pixel information for a given location is represented by \(I_{j}(\varvec{v})\).
PMVS makes no assumptions about the nature of the objects being reconstructed. It is likely that additional points are contained in X that comprise background or other non-plant material. Many such points will be removed by our level set approach; however, for computational efficiency many can be removed before reconstruction begins.
The point cloud is pre-filtered to remove obvious outliers; those points that differ greatly from the expected colour of the plant, or those that appear below the expected location of the plant. Two filters are applied, first a clipping plane positioned at the base of the plant is used to remove the majority of background points on the floor, container, etc. Second, colour filtering is achieved by examining the projected pixel values for every point, and removing those that do not appear green in hue. These filters are meant only as a conservative first pass, a more sensitive colour-based metric is used within the speed function during application of the level set method. The final filtered point cloud \(X^{\prime } \subseteq X\) is used in place of X for the remainder of the reconstruction process.
2.2 Point cloud clustering
The point cloud representation produced by PMVS contains no explicit surface description. Methods for the reconstruction of a surface mesh from a point cloud exist [17, 18]. Most, however, construct a single a surface describing the entire point cloud. Plants contain complex surface geometry that encourages the separation of leaves. We also wish to approach the more general problem of plant reconstruction, without assuming the connectivity or nature of the plant leaves is known. Instead, we model plant material as a series of small planar patches. Patch size is restricted to avoid fitting surfaces between nearby leaves, and to accurately model the curved nature of each leaf surface. The filtered point cloud is first clustered into small clusters of points using a radially-bounded nearest neighbour strategy [19]. Points are grouped with their nearest neighbours, as defined by a pre-set distance, and the method is extended to limit the potential size of each cluster. More formally, from the filtered cloud we obtain a set of clusters \({C_{k}}^{N_\mathrm{clus}}_{k=1}\) in which each cluster contains at least one point and all clusters are disjoint, so \(|C_{k}| > 0, \forall k\) and \(C_{k} \cap C_{l} = \emptyset , \forall k \ne l\).
This distance used for the nearest neighbour approach is dependent on the size and resolution of the model being captured. As PMVS (and laser scanning devices) usually output points with a consistent density, the distance parameter can be set once and then remain unchanged between experiments using the same image capture technique. Reducing this number will increase the number of planar sections fitted to the data, increasing accuracy at the cost of decreased algorithmic efficiency.
Our surface fitting approach begins with an approximation of the surface that will then be refined. A least-squares orthogonal regression plane is fitted to each cluster using singular value decomposition. This best fit plane minimises the orthogonal distance to each point, providing each cluster with a centre point \(\varvec{c}\), a normal vector \(\varvec{n}\), and an orthogonal vector \(\varvec{x}\) indicating the rotation about the normal. The vector \(\varvec{x}\) is aligned along the major-principle axis of the point within the cluster. We then define a set of orthographic projection functions that project individual world points into each cluster plane, \(\{\mathcal {C}_{k}(\varvec{w}): \mathbb {R}^{3}\rightarrow \mathbb {R}^{2}\}^{N_\mathrm{clus}}_{k=1}\), where \(\mathcal {C}_{k}\) represents the projection into plane k (i.e. the plane associated with cluster \(C_{k}\)). We say that points projected onto any plane now occupy planar co-ordinates. Any such point, denoted by the 2D vector \(\varvec{p}\), can be projected back into world co-ordinates by the set of functions \(\{\mathcal {W}_{k}(\varvec{p}): \mathbb {R}^{2}\rightarrow \mathbb {R}^{3}\}^{N_\mathrm{clus}}_{k=1}\).
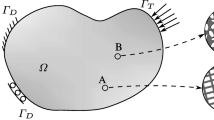
An overview of the geometrical co-ordinate systems used within our reconstruction framework. The model is represented in world co-ordinates, the perspective projection \(\mathcal {V}_{j}\) maps points in world co-ordinates into any given camera view i. The orthogonal projection \(\mathcal {C}_{k}\) maps points from world co-ordinates into any given surface patch k, which is projected back using \(\mathcal {W}_{k}\)
The orthogonal projection in \(\mathcal {C}_{k}\) has the effect of flattening the points in each cluster to lie on their best fit plane, reducing any noise in individual points, and reducing the surface fitting algorithm to a 2D problem. Point and mesh surfaces generated on a cluster plane will have an associated world position that can be output as a final 3D model. An overview of the geometric projections in use within our reconstruction approach can be seen in Fig. 1.
2.3 Surface estimation
An initial surface estimate is constructed by calculating the \(\alpha \)-shape of the set of 2D points in planar co-ordinates. An \(\alpha \)-shape is a generalisation of the convex hull for a set of points, and is closely related to the commonly used Delaunay triangulation. For the incomplete leaf surfaces that exist within the input cloud, the Delaunay triangulation and convex hull represent an over-simplification of the complex boundary topology of the clusters. For a point set S, Edelsbrunner [20] defines the concept of a generalised disk of radius \(1/\alpha \), with an edge between two points in S being included in the alpha shape if both points like on the boundary of the generalised disk, and that disk contains the entire point set. The set of \(\alpha \)-shapes, produced when varying alpha, represent a triangulation of each surface at varying levels of detail. In this work, a negative value of \(\alpha \) is used, with larger negative values removing larger edges or faces. The \(\alpha \) value can be tuned for a given data set, to preserve the shape of the boundary of each reconstructed point set.
2.4 Boundary optimisation
The \(\alpha \)-shapes computed over each cluster form an initial estimate of the location and shape of the plant surface. The challenging nature of plant datasets in multi-view reconstruction means that in many instances the initial point cloud estimate will be inaccurate or incomplete. The initial surface boundaries based on these points will require further optimisation to adequately reflect the true shape of each leaf surface. Missing leaf surfaces should be reconstructed, and overlapping shapes should be optimised to meet at a single boundary. Many methods, such as active contours [21], parameterise the boundary shape before attempting this optimisation. However, such approaches are ill suited to the complex boundary conditions produced by \(\alpha \)-shapes. For any value of \(\alpha < 0\), the surface may contain holes or disjoint sections, and as such many surfaces will change topology during any boundary optimisation process.
Tracking of such complex boundaries can be achieved using the level set method [22, 23]. The method defines a 3D function \(\varphi \) that intersects the cluster plane, with a single level set being initialised for each surface patch. \(\varphi \) is represented as a signed distance function, initialised such that negative values lie within our \(\alpha \)-shape boundary, and positive values occur outside. Thus, the boundary itself is defined as the set of all points in \(\varphi \) that intersect the cluster plane, given as:
A speed function determines the rate of change of \(\varphi \). It may be based on both global and local parameters, and will act to grow or shrink the boundary \(\Gamma \) as necessary to fit the underlying data. The change in \(\varphi \), based on a speed function v, is defined as
where \(\Delta \varphi \) is the gradient of the level set function at a given point, which we calculate through Godunov’s upwinding scheme. The speed function is defined as
where \(v_\mathrm{curve}\) is a measure of the local curvature, calculated using a central finite difference approximation
The curvature term encourages the boundary of the level set to remain smooth. The weighting \(\omega \) is required to prevent curvature from dictating the movement of the front, in cases where the boundary is already sufficiently smooth.
The image term, \(v_\mathrm{image}\), references colour information in the input images to ascertain whether the projection of the planar surface lies over regions with a high likelihood of containing leaf material. To achieve this, the function \(\varphi \) is discretized and uses the planar co-ordinate system, each planar point \(\varvec{p}\) maps to a position on \(\varphi \), and any point on \(\varphi \) will have an associated planar position. By performing consecutive projections, we are able to examine the relevant location in any image of a cluster plane position. Such a projection is given as \((\mathcal {V}_{i} \circ \mathcal {W}_{k})(\varvec{p}):\mathbb {R}^{2}\rightarrow \mathbb {R}^{2}\), where k is the cluster index, and i is the camera index. Not every image will provide a helpful view of every cluster, they may be out of the camera’s field of view, or seen at an oblique angle. One reference view is chosen from which to obtain colour information, as follows. We choose a reference image \(I_{R} \in I\) that represents a calculated “best view” of a planar surface. Selection of the reference view begins by projecting each cluster into each camera view. Only the interiors (triangular faces) of each \(\alpha \)-shape are projected using a scan-line rasterisation algorithm. Attached to each projected position is a z depth, calculated as the third component output from the function \(\mathcal {C}_{i}(\varvec{w})\) when using homogenous co-ordinates. This z depth represents the distance that the projected point lies from the camera’s image plane, and can be used to sort clusters that project onto the same location. Projections with the lowest z value are seen in front of, so occlude, those with higher z values.
The projection locations and z depths for all clusters are analysed using a series of z-buffer data structures, one z-buffer associated with each input image. We define the z-buffers as a set \(\{Z_{i}\}^{N_\mathrm{cam}}_{i=0}\), where each buffer contains pixel locations in camera co-ordinates that map directly to the corresponding image. For each image location, any cluster that can be seen in (i.e. projects onto) that point is recorded in the z-buffer. A given position \(Z_{i}(\varvec{v})\) contains a depth sorted list of all clusters that project into that camera co-ordinate, i.e. \(Z_{i}(\varvec{v}) = (C_{0}, \ldots , C_{n})\).
It is desirable to select camera views that contain as little interference between clusters as possible. For a given z-buffer j, and a given cluster i, we can calculate the following measure:
The clear pixel count represents a measure of the number of pixels each cluster projects into for a given image. This value reflects both the proximity of the cluster to the camera plane, and the angle of incidence between the camera view and the cluster plane. The clear pixel counts for all projections of a given cluster i are normalised to the range [0, 1]. This measure does not include pixel positions shared by other clusters, to avoid heavily occluded views affecting the normalised value. The amount of occlusion for each cluster i, in a given z-buffer j is calculated as:
where \(Z_{i}(\varvec{v})_{(k)}\) is the \(k\mathrm{th}\) ordered element of \(Z_{j}(\varvec{v})\). \(\mathcal {V}^\mathrm{occluded}_{j}(i)\) can be read as “the percentage of cluster i that projects into z-buffer j behind at least one other cluster.” Similarly, \(\mathcal {V}^\mathrm{occluding}_{j}(i)\) can be read as “the percentage of cluster i that projects into z-buffer j in front of at least one other cluster.” Thus, a combination of normalised clear pixel count, occlusion and occluding percentages can be used to sort images in terms of view quality. A reference image, \(I_{R}\), is chosen where
Penalising views that present occlusion with respect to each surface will help ensure that self-occlusion is prevented from affecting the reconstruction accuracy, only a single view of each surface needs to have an unobscured view for reconstruction of that patch to be successful.
When referencing pixel values using the image \(I_{R}\), we use a normalised green value to measure the likelihood of leaf material existing at that location,
We can assume that normalised green values will be higher in pixels containing leaf material, and lower in pixels containing background. Where lighting conditions remain consistent over an image set, we can also assume that distribution of normalised green values are the same over the each image in I. However, between different image sets we cannot assume that the properties of the normalised green values are known. These properties must be ascertained before \(\mathcal {N}_{i}\) can be used to contribute to the \(v_\mathrm{image}\) term in the speed function. We sample from all images those pixels that are projected into by the \(\alpha \)-shapes, and use Rosin’s unimodal thresholding approach [24] to threshold below the normalised green peak that is observed. Using this threshold, the mean and standard deviation of the peak are calculated, and used to produce an image speed function centred around the calculated threshold t, with a spread based on the standard deviation of the peak:

where t is the threshold calculated using Rosin’s method, and \(\sigma \) is the standard deviation of the \(\mathcal {N}_{j}\) peak. A width of \(2\sigma \) was chosen as a value that characterises the spread of the normalised green values.
The final component of the speed function, \(v_\mathrm{inter}\), works to reshape each surface based on the location and shape of nearby clusters. As each cluster may have different normal orientations, it is challenging to calculate their 3D intersections in terms of 2D positions in planar co-ordinates. Indeed, two nearby clusters that could be considered as overlapping, may not intersect in world co-ordinates. Instead we project each planar position into \(I_{R}\), and examine the interactions in the 2D camera co-ordinate system.
Any overlapping projections are calculated by maintaining z-buffers that update as each region reshapes. The function \(v_\mathrm{inter}\) is calculated such that each cluster in \(Z_{j}(x)\) is penalised except for the front-most cluster. Thus, for a cluster i, the function is calculated as:
where p is a small negative value such that the level set boundary \(\Gamma \) shrinks at this location. Note that the subtraction of \(v_\mathrm{image}\) results in the image component being ignored where clusters are occluded.
The complete speed function is used to update each discrete position on the level set function \(\varphi \). This process must be repeated until each cluster boundary has reshaped to adequately fit the underlying image data. The speed function will slow significantly as the boundary approaches an optimal shape. Where a level set boundary no longer moves with respect to the reference image (does not alter the number of projected pixels), we mark this cluster as complete and discontinue level set iterations. Any level sets that do not slow significantly will continue until a maximum time is elapsed, a parameter that can be set by the user. We typically use a value of 100–200 iterations as a compromise between computational efficiency and offering each level set adequate time to optimise.
2.5 Model output
Once all clusters have been iterated sufficiently, each surface triangulation must be re-computed. The level set function provides a known boundary that was not available during the original surface estimation. This can be used to drive a more accurate meshing approach that will preserve the contours of each shape. We use constrained Delaunay triangulation for this task [25]. A constrained triangulation will account for complex boundary shape when producing a mesh from a series of points; however, it will not over-simplify the boundary by fitting surfaces across concave sections, and can retain holes in the surface if required. Points are sampled from the boundary of each surface, and a constrained triangulation is fitted. This process will automatically generate additional points, where required, within the shape itself. As each point in the new triangulation exists in planar co-ordinates, they can be easily back-projected into world co-ordinates to be output in a 3D mesh format.
3 Experimental results
In this section, we present results obtained when applying our reconstruction approach to multiple views of single plants. Verification of our approach is achieved using a novel virtual dataset, in which a model rice plant is rendered from multiple viewpoints to generate artificial colour images, which are then treated in the same way as a real-world image set. This approach allows the reconstructed plant to be directly compared to the artificial target object, an impossible prospect when working with real-life plants, as no such ground truth can exist.
We have tested our reconstruction methods on datasets obtained from real rice and wheat plants, as well as an on real images of an artificial plant that exhibits a very different architecture. Images were captured using DSLR cameras with 35mm lenses, at 8 megapixel resolution. The number, and nature of the images were left to the user to decide given the subject in question, though we recommend more than 30 images surrounding the subject for a single plant. For the rice and wheat datasets, a single moving camera was used, and no special consideration was given to the environment in which the plants were imaged, beyond avoiding large areas of green colour in the background. The rice dataset was captured in an indoor environment, the wheat in a glass house. These environments provide complex backgrounds, which raise additional challenges, but the plants can still be reconstructed using our methods. The artificial plant was captured using three fixed-camera installations, and the plant rotated using a turntable. In this installation the turntable was rotated by hand in approximately 10–20\(^{\circ }\) increments.
Reconstruction of rice, wheat and the artificial plant images. (Top row) Sample images of the rice, wheat and artificial plant datasets. (Middle row) Meshed reconstructions of each plant surface using our approach. (Bottom row) Coloured representations of each plant once segmented using spectral clustering
In our experience a fixed-camera installation using a turntable often provides more reliable reconstructions than a moving camera installation. It is challenging to determine the best set of images for a given plant using a moving camera, particularly in an environment where other obstacles restrict the positions from which images can be captured. The lack of a robust protocol for image capture can lead to images being poorly distributed around a plant, missing some sections and increasing noise. The lack of background texture in the turntable installation information usually reduces the time required to capture the initial point cloud, where time is not spent reconstructing unnecessary background pixels that will simply be discarded later in the process. With no background, however, a textured target is required to ensure accurate calibration. This adds a further requirement that each camera view must see a sufficient proportion of the calibration target, meaning that as the height of the camera position is increased, the angle of view must also be increased. For taller plants this might mean a lack of adequate views of the uppermost leaves, and poor reconstructions in those areas. We anticipate that an automated turntable would solve this problem, where calibration could be accurately performed before reconstruction, and no textured target would be required once the plants were being captured.
Boundary refinement using the level set method. (Top left) An initial surface estimate of a section of the wheat dataset. (Top middle) A refined version of the wheat model after a level set was applied to each patch. (Bottom left) An initial surface estimate of a section of the rice dataset. (Bottom middle) A refined version of the rice model after a level set was applied to each patch. (Top right) Two example patches, viewed from the same position as the reference image \(I_{R}\). (Bottom right) A different orientation of the same two patches
Figure 2 shows the result of applying our reconstruction approach to the three image sets containing wheat, rice and the artificial plant. Quantitative evaluation of the effectiveness of any 3D shoot reconstruction is challenging due to a lack of ground truth models for comparison. Here we offer a qualitative evaluation of the benefits and shortcomings of our approach using these plants, followed by a quantitative evaluation using the virtual rice dataset.
Results on all three datasets showed that the initial surface estimate, obtained by calculating an \(\alpha \)-shape over each cluster, will naturally reproduce any flaws present in the PMVS point cloud. Most notable are the lack of point information in areas of poor texture, and noise perpendicular to the leaf surface, where depth has not been adequately resolved. These issues can be caused by the heavy self-occlusion observed in more dense plants or canopies, but are often caused in even simple datasets by a lack of image features in the centre of leaves. The artificial plant contains much larger leaves, however, texture is generally sufficient to provide a reliable set of points over each leaf surface.
Depth noise is significantly reduced by the use of best fit planes over small clusters, where all points are projected onto a single surface. However, the boundary of each surface is a function of the parameters used to create the \(\alpha \)-shape, and the quality of the underlying data. As such, we can expect the \(\alpha \)-shape boundaries to be a poor representation of the true leaf shape. With this in mind, we would characterise a successful reconstruction as one that significantly improves upon the initial surface estimate, through the optimisation of the each surface boundary.
Notable characteristics of the \(\alpha \)-shape boundaries in both datasets are significant overlap between neighbouring clusters, and frequent missing surface sections (Fig. 3). Figure 3 also shows the refined boundaries after the level set method has been applied, in which missing sections are filled, and overlapping surfaces have been reduced. The results in Fig. 3 are representative of the results over all three datasets.
(Top left) The original rice plant model, based on the plant reconstructed in Fig. 3. Vertices are coloured based on their mm distance to the nearest point on the reconstruction. (Bottom left) Histogram of smallest distances from each vertex on the model to vertices on the reconstruction. (Top right) The reconstruction produced by our approach. Vertices are coloured based on their mm distance to the nearest point on the original model. (Bottom right) Histogram of smallest distances from each vertex on the reconstruction to vertices on the model
While the refined surfaces represent an improvement over both the initial point cloud, and the initial \(\alpha \)-shape surface, there are still notable areas for improvement. By treating each section of leaf as an individually orientated plane, each plane orientation is susceptible to the error within the input cloud. Since each boundary is refined from one reference view, incorrect orientation of the best fit plane might cause the surface boundary to be incorrectly aligned with the image, or neighbouring clusters. Consider Fig. 3 (right), in which two patches have been reconstructed in close proximity. When viewed from the reference view in which boundary refinement occurred, the boundaries of neighbouring patches are in good agreement. A rotated view of the same surfaces, however, shows that misaligned normal orientation can lead to gaps between neighbouring surfaces. Conversely, if the right-hand image had been chosen as \(I_{R}\), the level set equation would increase the size of both boundaries, and overlap would be observed in the left hand view.
In reality, for many clusters with very similar orientations these gaps will be negligible; as the clusters are limited in size, the distance between neighbouring plane orientations will be small, and the resulting gaps between boundaries will also be small. We have quantified the low level of discrepancy between an input model and the reconstruction below. We anticipate that further work on smoothing the normal orientations of neighbouring clusters or merging neighbouring clusters into a single curved leaf model will continue to improve results in this regard: this will be a focus of upcoming research.
An additional dataset was created based on the plant used in the rice dataset. The rice plant was first manually captured and modelled using the point cloud created by PMVS, and 3D graphics software [26, 27]. This is a time consuming and subjective process, and should not be viewed as a suitable alternative to automatic reconstruction. However, it is possible to produce an easily quantifiable ground truth model that can be used as a target for automated reconstruction. This virtual plant was textured and coloured to emulate the original plant leaves. Finally, 40 distinct camera views of the model were rendered, simulating an image capture system moving around a static plant. The resulting dataset can then be reconstructed in the same manner as real-world data, while retaining the ability to compare the reconstruction with the original virtual plant, in particular keeping the same co-ordinate system and scale. The original model, and our reconstruction can be seen in Fig. 4.
To quantify the similarity between the original model and the reconstruction, we use the Hausdorff distance, the greatest distance from any point on either mesh, to the nearest point on the other. This concept is extended in [28] to include a measure of the mean distance between two meshes.
A visual representation of these measures can be seen in Fig. 4, in which each vertex is coloured based on the distance to the nearest point on the opposing mesh. This provides a visual clue as to our algorithm performance. The arbitrary world units used within the reconstruction were converted into mm measurements through the use of a calibration target of known size.
The furthest distance between points on both meshes is \(\sim \)4.5 mm; however, the average distances between each mesh are significantly lower. The complete model is approximately 48 cm tall. These one-sided measurements provide additional information, by distinguishing between the distances in either direction. Increasing distance from the model plant to the reconstruction indicates areas of the model that have not been accurately reconstructed. This is most likely where missing points in the initial cloud and surface estimates are not adequately refined through the level set method. In this case, the low mean and maximum distances show that these regions have been reconstructed successfully. Indeed, 99 % of the vertices in the model are within 1.2 mm of the reconstructed model (Table 1).
In the other direction, higher distances from the reconstruction to the original model represent areas that have deviated from the true position of the plant. This could be caused by a number of factors, such as misalignment between the orientation of a surface plane and the original surface, or surface boundaries extending beyond the true boundary of the leaves, possibly due to occlusion. The maximum and mean distances for the reconstruction remain low, and show that the reconstruction is a good reflection of the true model.
The mean distance and RMS error for this single-sided measure is higher than the reverse, which we believe may represent current technical limit of our approach. The distances around the boundaries of many surfaces appear slightly higher than in the centre, where the level sets can over-extend the leaf edge. This is a limitation within the level set speed function, but for the distances observed this usually represents an increase of size, outwards, of less than a pixel on average when projected into the reference image. This sub-pixel accuracy is not resolved by the speed function of the level set method that we use. An immediate improvement could be observed by simply increasing the resolution of the input image set; however, this would add significant computational overhead.
How accuracy of our reconstruction approach on the virtual dataset varies with size of the surface patches. The segmentation radius determines the size of the clusters obtained during point cloud clustering. Hausdorff distance here is measured relative to the size of the virtual plant model, lower is better
Our approach begins by clustering points based on a segmentation radius. During our experiments we used a radius of 0.03 world units, which was determined empirically. Our experience suggests that the approach is robust to changes in this value; however, in an effort to justify this choice we have tested the reconstruction accuracy on our virtual dataset as this parameter is changed (Fig. 5). The segmentation radius is a primary factor in determining the size of the surface patches that are produced, so a value should be chosen that is appropriate for the size of the planar regions of the leaves. Very small surface patches will cause the curvature term to become dominant during the level set iteration step, increasing the distance error. Very large patches will over-simplify the plant structure, also increasing the error. Values of 0.02 and 0.03 are seen to be effective, but note that other values still produce an error measure that is a fraction of 1 % of the size of the model.
The performance of our approach is closely related to the size of the image set, and the size of the model being evaluated (the number of patches, and their size). For small datasets, reconstruction usually takes a matter of minutes. For complex datasets, particularly those with more than 50 input images, we can expect performance to decrease. Table 2 shows details and processing times for the datasets evaluated in this section. Tests were run on an Intel Core i7 3820 machine. The algorithms detailed here are suitable for GPU parallelisation in the future if further optimisation is required.
In their raw form, these models represent a flexible way of measuring higher level plant traits. Any plant feature that can be directly mapped to an equivalent feature in a 3D model, can be captured. Thus, these models can be used for plant size, surface area, distributions of leaf angles, etc. More advanced measures specific to some areas of plant phenotyping can also be measured, such as leaf area index that is often used in photosynthetic modelling. However, given the output of this technique is a 3D model only, which measures are used and what approach is used to measure them is left to the end-user’s discretion. The models are also well suited to surface-based modelling approaches such as ray tracing [11]. For our approach to be suited to more general plant phenotyping, it is necessary to extract lower level phenotypic information about each plant, such as number and angle of leaves. Obtaining such measurements reliably for a variety of plant species is a goal for future research; however, as a proof of concept we were eager to show that the patch-based system we have employed can in principle be used to power lower level phenotyping. A spectral clustering approach offers a robust way to cluster surface patches into contiguous blocks, often leaves. We use the normalised spectral clustering approach outlined in [29]. Spectral clustering operates on an undirected graph \(G = (V,E)\), where in this case each vertex represents a single surface patch in the plant model. Edges between patches are weighted based on the distance between their centre points, but distorted to favour those that are closer parallel to the orientation of a plane, rather than orthogonal to it. More formally
where \(d_p = \mathbf {n}_i \cdot (\mathbf {c}_j - \mathbf {c}_i)\) and \(d_o = \Vert (\mathbf {c}_j - d_p \mathbf {n}_i) - \mathbf {c}_i \Vert \) with \(\Vert \cdot \Vert \) being Euclidean distance in three dimensions, \(\mathbf {c}_i\) and \(\mathbf {c}_j\) the centres of the two patches, and \(\mathbf {n}_i\) the unit normal to patch i. To make weights symmetric, we set weight \(w_{i,j} = \min (w_{i \rightarrow j}, w_{j \rightarrow i})\).
The weighted adjacency matrix of G is the matrix \(W = (w_{i,j})_{i,j=1,\ldots ,n}\), which we convert into a k-nearest neighbour representation by setting all but the k-closest neighbours of each vertex to zero. From this matrix we can calculate the degree matrix D, a diagonal matrix with degrees along the diagonal calculated as \(d_{i} = \sum _{j=1}^{n}w_{i,j}\). Finally, the normalised laplacian matrix L can be calculated as:
The eigenvectors of L that correspond to the k smallest eigenvalues are clustered row-wise using k-means++ [30]. The clusters assigned to each row are then mapped directly to the surface patches, resulting in a final segmentation.
The results of our initial clustering approach can be seen in Fig. 2. When leaves are well defined there is often strong separation between groups of patches into either complete leaves, or large sections of the same leaf. Towards the bases of each plant as the boundaries become increasingly hard to distinguish, performance decreases. It should be noted, however, that this is far from a complete solution, and is meant only to demonstrate the possibility of our patch-based model being used to extract more complex phenotypic measurements. The number of clusters k is currently determined empirically, along with the standard deviations \(\sigma _p^2\) and \(\sigma _o^2\) used to calculate the distance between patches. Further research will explore the possibility of improving the segmentation of these models, including the automatic determination of an optimal k, and an improved distance metric that includes patches that include boundaries in close proximity.
4 Conclusions
The recovery of accurate 3D models of plants from colour images, and their associated phenotypic traits, is a challenging topic. Even single plants represent a crowded scene in the sense of [13], and reconstructing objects with this level of complexity is an active research area, both within and outside the field of Plant Phenotyping. Plants often contain high degrees of self-occlusion, with the level of occlusion varying greatly even within a species. Individual leaves are also hard to identify, often exhibiting similarity, and lacking sufficient texture for many of the reconstruction approaches that see widespread use. For these reasons many existing plant reconstruction techniques have focused on the properties of plants that can be easily identified, in particular their silhouettes. Silhouette-based approaches have proven robust when reconstructing smaller, less detailed plants; however, performance will often deteriorate in the presence of increasing occlusion, as a plant ages, or multiple plants are imaged together. In our approach, where each surface is seen clearly from at least one camera, effective reconstruction can be performed.
The approach presented here attempts to address these issues by developing each leaf segment individually, automatically selecting an image that is likely to contain the necessary information for reconstruction. In essence, the problem of occlusion is reduced by choosing an image that has a clear view of each target surface. The problem of low texture is addressed through detailed analysis of the colours present in the image. Avoiding the use of texture improves performance of this approach on plants, when compared to standard feature-correspondence methods. The level set method re-sizes and re-shapes each patch as necessary to maximise its consistency with the reference image, as well as the consistency between nearby patches that might overlap. By driving the reconstruction without regard for leaves or plant structure, the approach remains general, and is flexible enough to be applied to a wide variety of plant species with differing leaf shape and pose. In its current form the mesh representation produced provides a detailed model of the surface of a viewed plant that can be used in both modelling tasks and for shoot phenotyping.
This general approach, however, makes the calculation of some plant traits less intuitive. General measurements such as surface area or height are easily obtained, but more plant-specific traits such as leaf count and angle cannot easily be measured on a patch-based model. To address this issue we have demonstrated that a spectral clustering approach is well suited to the task of grouping neighbouring patches, thus extending this approach to whole leaves. We anticipate that further work on leaf segmentation will yield many more useful plant trait measurements, without the loss of species generality.
References
Paproki, A., Sirault, X., Berry, S., Furbank, R., Fripp, J.: A novel mesh processing based technique for 3d plant analysis. BMC Plant Biol. 12(1), 63 (2012)
Quan, L., Tan P., Zeng, G., Yuan, L., Wang, J., Bing Kang, S.: Image-based plant modelling. In: ACM Transactions on Graphics (TOG), vol. 25, pp. 599–604. ACM, New York(2006)
Alenyà, G., Dellen, B., Torras, C.: 3d modelling of leaves from color and tof data for robotized plant measuring. In: 2011 IEEE International Conference on Robotics and Automation (ICRA), pp. 3408–3414. IEEE, New York(2011)
Kumar, P., Cai, J., Miklavcic, S.: High-throughput 3d modelling of plants for phenotypic analysis. In: Proceedings of the 27th Conference on Image and Vision Computing New Zealand, pp. 301–306. ACM, New York (2012)
Cai, J., Miklavcic, S.: Automated extraction of three-dimensional cereal plant structures from two-dimensional orthographic images. IET Image Process. 6(6), 687–696 (2012)
Houle, D., Govindaraju, D.R., Omholt, S.: Phenomics: the next challenge. Nature Rev. Genet. 11(12), 855–866 (2010)
White, J.W., Andrade-Sanchez, P., Gore, M.A., Bronson, K.F., Coffelt, T.A., Conley, M.M., Feldmann, K.A., French, A.N., Heun, J.T., Hunsaker, D.J. et al.: Field-based phenomics for plant genetics research. Field Crops Res. 133, 101–112 (2012)
Ma, W., Zha, H., Liu, J., Zhang, X., Xiang, B.: Image-based plant modeling by knowing leaves from their apexes. In: 19th International Conference on Pattern Recognition, 2008. ICPR 2008, pp. 1–4. IEEE, New York (2008)
Clark, R.T., MacCurdy, R.B., Jung, J.K., Shaff, J.E., McCouch, S.R., Aneshansley, D.J., Kochian, L.V.: Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol. 156(2), 455–465 (2011)
Kutulakos, K.N., Seitz, S.M.: A theory of shape by space carving. Int. J. Comput. Vis. 38(3), 199–218 (2000)
Song, Q., Zhang, G., Zhu, X.G.: Optimal crop canopy architecture to maximise canopy photosynthetic co2 uptake under elevated co2-a theoretical study using a mechanistic model of canopy photosynthesis. Funct. Plant Biol. 40(2), 108–124 (2013)
Pound, M.P., French, A.P., Murchie, E.H., Pridmore, T.P.: Automated recovery of three-dimensional models of plant shoots from multiple color images. Plant Physiol. 166(4), 1688–1698 (2014)
Furukawa, Y., Ponce, J.: Accurate, dense, and robust multiview stereopsis. IEEE Trans. Pattern Anal. Mach. Intell. 32(8), 1362–1376 (2010)
Wu, C.: Visualsfm: a visual structure from motion system (2011)
Lowe, D.G.: Object recognition from local scale-invariant features. In: The Proceedings of the Seventh IEEE International Conference on Computer Vision, 1999, vol. 2, pp. 1150–1157. IEEE, New York (1999)
Hartley, R., Zisserman, A.: Multiple View Geometry in Computer Vision. Cambridge University Press, Cambridge (2003)
Carr, J.C., Beatson, R.K., Cherrie, J.B., Mitchell, T.J., Richard Fright, W., McCallum, B.C., Evans, T.R.: Reconstruction and representation of 3d objects with radial basis functions. In: Proceedings of the 28th Annual Conference on Computer Graphics and Interactive Techniques, pp. 67–76. ACM, New York (2001)
Kazhdan, M., Bolitho, M., Hoppe, H.: Poisson surface reconstruction. In: Proceedings of the Fourth Eurographics Symposium on Geometry Processing, vol. 7 (2006)
Klasing, K., Wollherr, D., Buss, M.: A clustering method for efficient segmentation of 3d laser data. In: ICRA, pp. 4043–4048 (2008)
Edelsbrunner, H., Kirkpatrick, D., Seidel, R.: On the shape of a set of points in the plane. IEEE Trans. Inf. Theory 29(4), 551–559 (1983)
Kass, M., Witkin, A., Terzopoulos, D.: Snakes: active contour models. Int. J. Comput. Vis. 1(4), 321–331 (1988)
Osher, S., Sethian, J.A.: Fronts propagating with curvature-dependent speed: algorithms based on Hamilton–Jacobi formulations. J. Comput. Phys. 79(1), 12–49 (1988)
Sethian, J.A.: Level Set Methods and Fast Marching Methods: Evolving Interfaces in Computational Geometry, Fluid Mechanics, Computer Vision, and Materials Science, vol. 3. Cambridge University Press, Cambridge (1999)
Rosin, P.L.: Unimodal thresholding. Pattern Recognit. 34(11), 2083–2096 (2001)
Shewchuk, J.R.: Delaunay refinement algorithms for triangular mesh generation. Comput. Geom. 22(1), 21–74 (2002)
Sc pixelmachine srl topogun, v2.0. http://www.topogun.com
Blender foundation: Blender, v2.69. http://www.blender.org
Cignoni, P., Rocchini, C., Scopigno, R.: Metro: measuring error on simplified surfaces. In: Computer Graphics Forum, vol. 17, pp. 167–174. Wiley Online Library, New York (1998)
Shi, J., Malik, J.: Normalized cuts and image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 22(8), 888–905 (2000)
Arthur, D., Vassilvitskii, S.: k-means++: the advantages of careful seeding. In: Proceedings of the Eighteenth Annual ACM-SIAM Symposium on Discrete Algorithms, pp. 1027–1035. Society for Industrial and Applied Mathematics (2007)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Pound, M.P., French, A.P., Fozard, J.A. et al. A patch-based approach to 3D plant shoot phenotyping. Machine Vision and Applications 27, 767–779 (2016). https://doi.org/10.1007/s00138-016-0756-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00138-016-0756-8