Abstract
Soil sorption, described as logKOC (the logarithm of the soil/water partition coefficient normalized to organic carbon), was modeled using the augmented multivariate image analysis applied to quantitative structure–property relationship method for a series of 11 carboxylic acid herbicides. The statistical model was found to be highly predictive and reliable to estimate logKOC of other persistent organic pollutants in the soil, which are analogues of the carboxylic acids used in the QSPR model. The QSPR model derived from images corresponding to the chemical structures of the 11 herbicides is superior to the uniparameter model based on the octanol/water partition coefficient (logP) and, in addition, a pattern recognition model was built using principal component analysis. This model allowed clustering and separating compounds with low/moderate soil sorption from those with moderate/high soil sorption (compounds with the aryloxy function) using the second principal component.
Similar content being viewed by others
Carboxylic acid herbicides, such as 2,4-dichlorophenoxyacetic acid (2,4-D), are widely used in the control of broadleaf weeds, by complexing with the TIR1 ubiquitin ligase enzyme, then controlling plant growth and development (Tan et al. 2007). However, many herbicides are also persistent organic pollutants (POPs), which persist for very long periods of time in the environment and consequently may accumulate to a high level in the food chain, causing toxic effects like problems in reproduction, development and immunological functions (Corsonlini et al. 2005; Domingo 2004; Giesy et al. 1994; Kavlock et al. 1996; Kelce et al. 1995; Ratcliffe 1967, 1970). Consequently, developers of new herbicides should consider their environmental risk in addition to efficacy.
Quantitative structure–property relationship methods can be used to understand the effect of structural changes in herbicide molecules and to predict their properties, such as bioactivity and toxicity. The soil/water partition coefficient normalized to organic carbon (KOC) can be used to determine the environmental fate and persistence of POPs. It is usually linearly regressed against KOW values, the octanol/water partition coefficient, which can be either experimentally determined or estimated using calculations based on contributions from molecular fragments. However, such a correlation is not consistent for some classes of herbicides, like triazine and acetanilide-type herbicides (Freitas et al. 2014). Thus, alternative QSPR methods to better encode chemical structures using more representative molecular descriptors are required.
While 3D-QSAR/QSPR methods have been widely used to model the bioactivities and diverse properties of molecules, 2D approaches have not shown to be inferior to three-dimensional descriptors in many cases (Brown and Martin 1997; Estrada et al. 2001). Indeed, the three-dimensional structure of 2,4-D and analogs has not been considered fundamental to be taken into account in QSPR studies (Freitas and Ramalho 2013). Thus, the aug-MIA-QSPR method (Nunes and Freitas 2013), which is based on 2D drawings of chemical structures containing colored spheres representing different atoms with varying Van der Waals radii, is expected to correlate appropriately the molecular descriptors with the property to be investigated. Indeed, such an approach has been successful in describing the phytotoxicities of benzoxazinone herbicides and related compounds on problematic weeds (Freitas et al. 2013), and it is proposed here to model the logKOC and logP values of a series carboxylic acid herbicides.
Materials and Methods
Experimental values of logKOC and logP were obtained from the literature for a series of 11 carboxylic acid herbicides (Mackay et al. 1997) and their chemical structures were drawn using the GaussView program. The aug-MIA-QSPR has been described in details elsewhere (Nunes and Freitas 2013); thus, only a brief description is given here. Spheres representing atoms were drawn proportionally to the respective Van der Waals radii (the covalent Van der Waals radii were scaled to 75 %) and each atom type had a different color, whose numerical value (according to the RGB color system) was proportional to the atomic electronegativity. Each drawing (chemical structure) was saved as an individual bitmap file using the Paint application of the Microsoft Windows. It is worth mentioning that images should be drawn systematically, that is the first molecule was drawn with the congruent substructure (the COOH group) fixed in a given position of the GaussView workspace. To make the remaining molecules superimposed with the first (2D alignment), the carboxyl group was retained and the remaining organic chains in the other molecules replaced that of the first molecule, and then they were subsequently copied and pasted in the Paint application of the Microsoft Windows to be saved as bitmaps. The images were numerically converted according to the RGB color system using the Matlab program. The files were grouped to obtain a x × y × z three-way array, in which x corresponds to the number of samples (compounds), while y and z correspond to the coordinates of the pixels composing each image, whose variance explains the changes in the y block (the soil sorption column vector). The superposition of the 11 images is shown in Fig. 1; the structural changes explain the variance in the logKOC data, which is required to study the quantitative structure–property relationship. The 3D array was unfolded to a 2D matrix [x × (y × z)] and then regressed against the logKOC data using partial least squares (PLS) regression. A similar procedure was developed for the correlation with the logP data.
Results and Discussion
Frequently, the soil sorption (logKOC) of herbicides is indirectly estimated using a well-known relationship with the octanol–water partition coefficient (logP). However, such a correlation does not exist for the carboxylic acid herbicides of Table 1. The determination coefficient (r 2) found between logKOC and logP was negligible (0.35) and, after removing three apparent outliers (compounds 1, 3 and 7, with high standard deviation between experimental and calculated values), r 2 improved insufficiently to 0.50. Thus, a more complex QSPR model is required to encode appropriately the relationship between chemical structure and logKOC.
The aug-MIA-QSPR model obtained from the images of 10 herbicides (compound 7 was removed because it was identified as an outlier) gave a significant correlation between descriptors of chemical structures and logKOC (recommended r 2 > 0.8), according to the statistical data of Table 2 and low root mean square errors (RMSE) between experimental and calculated values. The aug-MIA-QSPR model was validated using leave-one-out cross-validation (LOOCV), giving a recommended q 2 above 0.5. The relatively high residual in the LOOCV for compound 1 is due to its uniqueness as aliphatic compound within the series. The y-randomization test identifies whether or not a model retains a statistically high r 2 value after shuffling the column vector containing the logKOC data and keeping intact the descriptors matrix; if a correlation between chemical structure and logKOC really exists, then a correlation with the randomized data is expected to be poor. This was confirmed by the parameter \( ^{\text{c}} r^{ 2}_{\text{P}} \) > 0.5 (Mitra et al. 2010), described as \( ^{\text{c}} r^{ 2}_{\text{P}} \) = r × (r 2 − r 2 y-rand )1/2.
Table 3 and Fig. 2 show that, in addition to encode logKOC, the aug-MIA descriptors obtained from the images of chemical structures of the carboxylic acid herbicides also describe the logP data. Such a description is not achieved using some popular methods to calculate logP from fragment-based chemical structure. For instance, the correlation between experimental and calculated logP values obtained from the Percepta module of the ACD/Labs program gave r 2 of 0.551 using all 11 herbicides and 0.515 using 9 herbicides of Table 1 (1 and 3 removed). The high residual in the LOOCV for compound 4 is due to its uniqueness as heterocyclic compound within the series. Thus, aug-MIA descriptors for the series of carboxylic acid herbicides studied can be used to predict both logKOC and logP of congeneric herbicides.
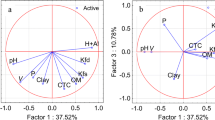
The aug-MIA descriptors were also used to build a classification model using principal component analysis (PCA, Fig. 3). The first principal component (PC1) separated the single aliphatic compound of the series (at left in PC1) and compounds with larger carbon chain (at right in PC1) from the remaining compounds. Positive scores in PC2 (at the top in the scores plot) indicate carboxylic acid herbicides with low and moderate soil sorption; negative scores in PC2 (at the bottom in the scores plot) indicate herbicides with moderate and high soil sorption. Overall, the non-aromatic compound 1 is moderately sorbed, while carboxylic acid herbicides containing an additional aryloxy function in the same chain of the carboxylic group have moderate/high soil sorption.
Images representing chemical structures of some carboxylic acid herbicides encode the logKOC and logP properties. Both regression (quantitative) and pattern recognition (qualitative) models were obtained using aug-MIA descriptors, which can then be used to predict the profile of carboxylic acid herbicides with respect to their soil sorption and hydrophobicity. Herbicides containing aromatic groups without the ether function in the same carbon chain of the carboxylic group tend to be promising herbicides with low soil sorption and, thus, corresponding analogs may drive the development of new carboxylic acid herbicides with decreased environmental hazard.
References
Brown RD, Martin YC (1997) The information content of 2D and 3D structural descriptors relevant to ligand-receptor binding. J Chem Inf Comput Sci 37:1–9
Corsonlini S, Ademollo N, Romeo T, Greco S, Focardi S (2005) Persistent organic pollutants in edible fish: a human and environmental health problem. Microchem J 79:115–123
Domingo JL (2004) Human exposure to polybrominated diphenyl ethers through the diet. J Chromatogr A 1054:321–326
Estrada E, Molina E, Perdomo-López I (2001) Can 3D structural parameters be predicted from 2D (topological) molecular descriptors? J Chem Inf Comput Sci 41:1015–1021
Freitas MP, Ramalho TC (2013) Employing conformational analysis in the molecular modeling of agrochemicals: insights on QSAR parameters of 2,4-D. Ciênc Agrotechnol 37:485–494
Freitas MR, Matias SVBG, Macedo RLG, Freitas MP, Venturin N (2013) Augmented multivariate image analysis applied to quantitative structure–activity relationship modeling of the phytotoxicities of benzoxazinone herbicides and related compounds on problematic weeds. J Agric Food Chem 61:8499–8503
Freitas MR, Matias SVBG, Macedo RLG, Freitas MP, Venturin N (2014) Three-parameter modeling of the soil sorption of acetanilide and triazine herbicide derivatives. Bull Environ Contam Toxicol 92:143–147
Giesy JP, Ludwig JP, Tillitt DE (1994) Deformities in birds of the Great-Lakes region assigning causality. Environ Sci Technol 28:A128–A135
Kavlock RJ, Daston GP, Derosa C, Fenner-Crisp P, Gray LE, Kaattari S, Lucier G, Luster M, Mac MJ, Maczka C, Miller R, Moore J, Rolland R, Scott G, Sheehan DM, Sinks T, Tilson HA (1996) Research needs for the risk assessment of health and environmental effects of endocrine disruptors: a report of the US EPA sponsored workshop. Environ Health Perspect 104:715–740
Kelce WR, Stone CR, Laws SC, Gray LE, Kemppainen JA, Wilson EM (1995) Persistent DDT metabolite P, P’-DDE is a potent androgen receptor antagonist. Nature 375:581–585
Mackay D, Shiu W-Y, Ma K-C (1997) Illustrated handbook of physical-chemical properties and environmental fate for organic chemicals. Lewis Publishers, New York
Mitra I, Saha A, Roy K (2010) Exploring quantitative structure–activity relationship studies of antioxidant phenolic compounds obtained from traditional Chinese medicinal plants. Mol Simul 36:1067–1079
Nunes CA, Freitas MP (2013) Introducing new dimensions in MIA-QSAR: a case for chemokine receptor inhibitors. Eur J Med Chem 62:297–300
Ratcliffe DA (1967) Decrease in eggshell weight in certain birds of prey. Nature 215:208–210
Ratcliffe DA (1970) Changes attributable to pesticides in egg breakage frequency and eggshell thickness in some British birds. J Appl Ecol 7:67–115
Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446:640–645
Acknowledgments
Authors are grateful to FAPEMIG, CNPq and CAPES for the financial support of this research, as well as for the fellowships.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Freitas, M.R., Freitas, M.P. & Macedo, R.L.G. Aug-MIA-QSPR Modeling of the Soil Sorption of Carboxylic Acid Herbicides. Bull Environ Contam Toxicol 93, 489–492 (2014). https://doi.org/10.1007/s00128-014-1356-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00128-014-1356-9