Abstract
Aim/hypothesis
Hepatic forkhead box q1 (FOXQ1) expression levels are regulated by nutritional and pathophysiological status. In this study we investigated the role of FOXQ1 in the regulation of hepatic gluconeogenesis.
Methods
We used multiple mouse and cell models to study the role of FOXQ1 in regulating expression of gluconeogenic genes, and cellular and hepatic glucose production.
Results
Expression of hepatic FOXQ1 was regulated by fasting in normal mice and was dysregulated in diabetic mice. Overexpression of FOXQ1 in primary hepatocytes inhibited expression of gluconeogenic genes and decreased cellular glucose output. Hepatic FOXQ1 rescue in db/db and high-fat diet-induced obese mice markedly decreased blood glucose level and improved glucose intolerance. In contrast, wild-type C57 mice with hepatic FOXQ1 deficiency displayed increased blood glucose levels and impaired glucose tolerance. Interestingly, studies into molecular mechanisms indicated that FOXQ1 interacts with FOXO1, thereby blocking FOXO1 activity on hepatic gluconeogenesis, preventing it from directly binding to insulin response elements mapped in the promoter region of gluconeogenic genes.
Conclusions/interpretation
FOXQ1 is a novel factor involved in regulating hepatic gluconeogenesis, and the decreased FOXQ1 expression in liver may contribute to the development of type 2 diabetes.
Similar content being viewed by others
Introduction
Hepatic gluconeogenesis is absolutely required for survival during prolonged fasting, however, abnormal elevation of hepatic glucose production contributes to fasting hyperglycaemia in diabetes [1]. Efforts to uncover the molecular mechanisms that control hepatic gluconeogenesis are crucial for the development of new strategies for the treatment of diabetes [2].
The expression of key gluconeogenic genes, Pck1 and G6pc, which encode phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase), respectively, is controlled at the transcriptional level by hormones, including insulin, glucagon and glucocorticoids [3, 4]. In contrast, insulin inhibits hepatic gluconeogenesis by negatively regulating transcriptional factors, including FOXO1 and PGC-1α [2, 5–10].
Forkhead box O1 (FOXO1 [also known as forkhead transcription factor, FKHR]) acts as a cardinal regulator of whole-body energy homeostasis [11]. FOXO1 contains highly conserved Akt phosphorylation sites (Thr24, Ser253 and Ser316). Phosphorylated FOXO1 induced by increased insulin levels is excluded from the nucleus, thereby decreasing its transcriptional activity [5, 12, 13]. In contrast, the decreased blood insulin levels during fasting promote FOXO1 nuclear localisation, where it collaborates with peroxisome proliferator-activated receptor, γ, co-activator 1 α (PGC-1α) to increase the expression of the key gluconeogenic genes, Pck1 and G6pc, via direct binding to insulin response elements (IREs) in their promoters [2, 11, 14, 15]. Hepatic FOXO1 deficiency in mice impairs fasting-induced gluconeogenesis, subsequently leading to lowered blood glucose level [8].
FOXQ1 (also known as hepatic nuclear factor-3 homolog-1, HFH1), a member of the FOXQ subfamily, is expressed in multiple tissues, including kidney, stomach, liver, bladder and other tissues [16–18]. FOXQ1 has a role in hair follicle development and FOXQ1 mutant mice have a silky shiny coat appearance [17]. Mice with FOXQ1 deficiency exhibit a lack of gastric acid secretion [19]. Accumulating evidence suggests that increased FOXQ1 expression is correlated with metastasis and poor prognosis for multiple human cancers, including breast cancer, colon cancer and lung cancer [20–23].
In this study, we investigate the role of FOXQ1 in the regulation of hepatic glucose metabolism.
Methods
Animals and experimental design
Male db/db mice and C57BL/6J mice at 8–9 weeks of age were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China) and housed and maintained in a 12-h light and dark photoperiods.
Mice were randomised and experimenters were blind to group assignment. db/db mice used for study were greater than 39 g in weight and had blood glucose levels over 15.1 mmol/l. Mice were injected with adenovirus containing green fluorescent protein (Ad-gfp), adenovirus expressing Foxq1 (Ad-Foxq1), short-hairpin RNA (shRNA) control adenovirus expressing shRNA against luciferase (Ad-shCtrl) or adenovirus expressing shRNA against Foxq1 (Ad-shFoxq1; see below for further methodology details). Then, 7–8 days after infection, mice were fasted for 6 h and their livers were collected for further analysis.
Preparation of expression plasmids and recombinant adenoviruses
The full-length mouse Foxq1 or Foxo1 gene was amplified by PCR from C57BL/6J mouse liver cDNA. The myc-tagged Foxq1 was cloned into pcDNA3.1(Invitrogen, Carlsbad, CA, USA). A flag-tag at the C-terminus of Foxo1 was constructed into pAd-Track-CMV (Addgene, Cambridge, MA, USA). Specific primers are shown in ESM Methods. Recombinant adenoviruses expressing Foxq1 or Foxo1 were generated as previously described [24].
The myc-haemagglutinin-(HA-)Foxo1 expression plasmid was a kind gift from D. Accili (Columbia University, New York, NY, USA).
RNA interference
shRNA adenoviruses were prepared according to previously described methods [24]. The sequences of small interfering RNA transcribed from Ad-shCtrl and Ad-shFoxq1 are shown in ESM Methods.
Quantitative real-time PCR
Total RNA was isolated from cells or pulverised liver using TRIzol (Invitrogen). Quantitative PCR (qPCR) was performed as previously described [25]. mRNA levels for specific genes were normalised to β-actin mRNA levels and expressed relative to their mRNA levels from control samples. Specific primers are shown in ESM Methods.
Western blot analysis
Western blot assays were performed as previously described [25] using antibodies specific for rabbit anti-PGC-1α (1:2000), rabbit anti-G6Pase (1:1000), mouse anti-Flag (1:2000), mouse anti-c-Myc (1:5000), rabbit anti-FOXO1 (1:2000) and mouse anti-β-actin (1:2000). A polyclonal antibody against FOXQ1 (1:700) was generated in our laboratory by immunising rats with glutathione S-transferase (GST)-full-length FOXQ1 fusion protein. See ESM methods for antibody details.
Cell culture and treatment
Primary mouse hepatocytes were isolated from the livers of male C57BL/6J mice (8 weeks old) and cultured as previously described [25]. Mouse hepatocytes were infected with indicated adenoviruses. Twenty-four h (for Ad-Foxq1 and Ad-gfp treatment) or 48 h (for Ad-shFoxq1 and Ad-shCtrl treatment) after infection, cells were cultured in serum-free medium and treated with or without dexamethasone (DEX; 1 μmol/l; Sigma-Aldrich, St. Louis, MO, USA) and forskolin (FSK; 10 μmol/l; Sigma-Aldrich) for 6 h prior to harvest for further analysis.
Analytical procedures and chemicals
Blood glucose values were determined using a glucometer (One Touch Ultra; LifeScan, Milpitas, CA, USA). Serum concentrations of triacylglycerol, cholesterol, NEFA, alanine transaminase (ALT) and aspartate aminotransferase (AST) were determined using an automated Monarch device ( Instrumentation Laboratory, Lexington, MA, USA). Serum insulin concentrations were determined by ELISA (R&D Systems, Minneapolis, MN, USA).
In vivo glucose-, insulin- and pyruvate tolerance tests
For in vivo infections, adenoviruses (Ad-gfp, Ad-Foxq1, Ad-shCtrl or Ad-shFoxq1) were delivered by tail vein injection into db/db, diet-induced obese mice (DIO) or control mice, both from a C57BL/6J mice. Five days after injection, mice were injected with d-glucose (1–2 g/kg) or pyruvate sodium (1–1.5 g/kg) via i.p. injection after 16 h fasting, and blood glucose levels were determined using a glucose monitor. For the insulin tolerance test (ITT), mice were injected with insulin (0.5–0.75 U/kg) via i.p. injection after 6 h fasting.
Luciferase reporter gene assay
Luciferase reporter gene was cotransfected into HepG2 cells, together with other expression plasmids, as indicated. A Renilla luciferase expression vector, pCMV-RL-TK (Promega, Madison, WI, USA), was used as an internal control to adjust for transfection efficiency. After 48 h , cells were harvested to assess luciferase activity using the Dual Luciferase Reporter Assay System (Promega). Relative luciferase activity was corrected for Renilla luciferase activity of pCMV-RL-TK and normalised to the control activity.
Electromobility shift assay
Electromobility shift assay (EMSA) was performed using the LightShift Chemiluminescent EMSA kit (Pierce, Appleton, WI, USA) as previously described [26]. Briefly, HEK293A cells were transfected with FOXO1, FOXO1+FOXQ1 and FOXQ1 expression plasmids. Nuclear proteins were extracted according to the manufacturer’s protocol (Pierce). Biotin-labelled, double-stranded oligonucleotides containing the wild-type IRE of the G6pc promoter were incubated with or without nuclear extracts. The protein–DNA complexes were detected with chemiluminescence reagent. Specific primers are shown in ESM Methods.
Glucose production assays
Primary mouse hepatocytes were infected with indicated adenovirus for 48 h and the glucose concentration in the medium was measured with a colorimetric glucose assay kit (Sigma-Aldrich).
Chromatin immunoprecipitation assay
Primary mouse hepatocytes were infected with indicated adenovirus for 24 h, then treated with RPMI 1640 medium plus DEX and FSK for 2 h. Cells were lysed and sonicated as previously described [27]. The protein–DNA complexes were immunoprecipitated with mouse IgG antibodies (control) or anti-FOXO1 antibody. The promoter region of G6pc containing the IRE was amplified by qPCR using specific primers shown in ESM Methods.
Co-immunoprecipitation experiments
Myc-tagged FOXQ1 and myc-HA-tagged FOXO1 or flag-tagged FOXO3 were transfected into HEK293A cells. Whole cell lysates, prepared after 36 h transfection, were incubated overnight with antibodies against FOXQ1 or Myc at 4°C followed by treatment with protein G agarose beads (GE Healthcare Life Sciences, Fairfield, CT, USA) for 2 h. The immunoprecipitates were immunoblotted with antibodies against FOXO1 or flag.
GST pull-down assay
GST-full-length FOXQ1 fusion protein was expressed in BL21 (DE3) strain of Escherichia coli using the pGEX-4T-1 vector (GE Healthcare Life Sciences). The GST fusion protein was purified with glutathione sepharose 4B (GE Healthcare Life Sciences). Myc-HA-tagged FOXO1 or flag-tagged FOXO1 fragments plasmids were transfected into HEK293A cells. Whole cell lysates were collected and mixed with glutathione sepharose-bound GST-FOXQ1 overnight at 4°C with gentle rocking. The glutathione sepharose beads were washed five times with lysis buffer, and immunoblotted using antibodies against FOXO1 or Flag (see ESM Methods for antibody details).
Statistical analysis
Data are presented as means ± SEM and were compared between or among groups by a two-tailed unpaired Student’s t test or by a one-way ANOVA, followed by a Fisher least significant difference test. A value of p < 0.05 was considered statistically significant.
Results
Hepatic Foxq1 gene expression is regulated by nutritional status and is dysregulated in diabetic and DIO mice
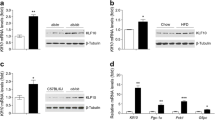
Multiple forkhead transcription factors, including FOXO1, FOXA2 and FOXC2, have been shown to be involved in glucose and lipid metabolism [28, 29]. However, whether and how the other FOX family members control systemic metabolism remains largely unclear. To identify potential FOX genes involved in dysfunctional hepatic glucose and lipid homeostasis in diabetes and obesity, we first systematically studied the expression of the FOX family members in the livers of db/db mice, a widely used type 2 diabetic model (ESM Table 1). Our qPCR and western blot results revealed that the expression levels of FOXQ1 were markedly decreased in the livers of db/db mice compared with db/+ control mice (Fig. 1a). In contrast, gluconeogenic genes, including Pgc-1α, G6pc and Pck1, showed an opposite expression pattern in db/db mice. In addition, we observed that hepatic Foxq1 mRNA levels were also reduced in high-fat DIO mice (from a C57BL/6J background) compared with mice fed a normal chow diet (Fig. 1b).
Hepatic FOXQ1 gene expression is regulated by nutritional status and dysregulated in diabetic and DIO mice. (a) qPCR analysis of Foxq1, Pgc-1α, G6pc and Pck1 mRNA expression, and western blot analysis of hepatic FOXQ1 expression in 8-week-old male mice. db/+ control mice, black bars; diabetic (db/db) mice, white bars. (b) Foxq1 mRNA expression in C57BL/6J mice fed a normal chow (control mice) or a high-fat diet (HFD) for 16 weeks (DIO mice). (c) Expression of Foxq1, Pgc-1α, G6pc and Pck1 mRNA, and protein expression of hepatic FOXQ1 and PGC-1α in 8-week-old male C57BL/6J mice fed ad libitum (black bar) or fasted for 24 h (white bar). n = 4 for all. Data are presented as means ± SEM. *p < 0.05, **p < 0.01
Next, we studied whether the expression of hepatic FOXQ1 could be regulated by nutritional status. Prolonged fasting (24 h) led to a decrease in mRNA and protein levels of FOXQ1 in the livers of wild-type C57BL/6J mice (Fig. 1c). However, genes involved in gluoneogenesis, including Pgc-1α, G6pc and Pck1, were markedly induced during prolonged fasting conditions (Fig. 1c). Additionally, short-term fasting (6 h) also decreased hepatic Foxq1 mRNA levels (ESM Fig. 1). Our data imply a strong correlation between FOXQ1 expression and glucose metabolism in the liver.
To identify potential triggers of FOXQ1 repression under fasting and pathophysiological conditions, primary hepatocytes were isolated and treated with FSK and DEX. FSK increases cAMP levels, thereby mimicking the effect of both glucagon and catecholamines on glucose production in hepatocytes. DEX mimics the effects of glucocorticoids, activating the gluconeogenic programme [3]. Consistent with the previous report, the treatment of cells with these hormones markedly induced expression of gluconeogenic genes (Fig. 2a), while this treatment significantly decreased the expression of Foxq1 (Fig. 2a). To further discern the effect of FSK and DEX on Foxq1 expression, we treated primary hepatocytes with these hormones, either individually or in combination. As a result, treatment with DEX alone significantly decreased FOXQ1 expression, while FSK alone had a minimal effect. Additionally, insulin did not significantly affect FOXQ1 expression (ESM Fig. 2a). These data suggest that glucocorticoids sufficiently regulate Foxq1 expression in a cell-autonomous manner.
FOXQ1 regulates the gluconeogenic programme in primary hepatocytes. (a) Quantitative PCR analysis showing mRNA levels of Foxq1, Pgc-1α, G6pc and Pck1 in mouse primary hepatocytes treated with or without DEX and FSK. (b) Western blot results showing FOXQ1-myc fusion protein (Myc) levels in mouse primary hepatocytes infected with Ad-gfp or Ad-Foxq1. (c) Quantitative PCR analysis showing the mRNA levels of Pgc-1α, G6pc and Pck1 in mouse primary hepatocytes infected with Ad-gfp or Ad-Foxq1 in the presence or absence of DEX and FSK. (d) Glucose production (glucose output) in primary hepatocytes treated as in (c). Results are presented as means ± SEM of triplicate experiments. *p < 0.05, **p < 0.01
To test the functional significance of FOXQ1 expression in the gluconeogenic programme, primary mouse hepatocytes were isolated and infected with Ad-Foxq1 expressing myc-tagged Foxq1. Western blot results indicated this adenovirus effectively induced expression of the FOXQ1 protein (Fig. 2b). Ad-Foxq1 treatment significantly inhibited the expression of gluconeogenic genes in primary hepatocytes in the presence of FSK and DEX (Fig. 2c). Accordingly, the forced expression of FOXQ1 markedly decreased glucose production in hepatocytes exposed to FSK and DEX (Fig. 2d). To further explore the role of FOXQ1 in mediating effects of DEX and FSK on gluconeogenic genes, we treated primary hepatocytes with Ad-shFoxq1 expressing FOXQ1-specific shRNA. Knockdown of FOXQ1 increased expression of gluconeogenic genes in the absence or presence of DEX and FSK (ESM Fig. 2b,c). These results indicate that FOXQ1 may inhibit the gluconeogenic programme in mouse primary hepatocytes.
Hepatic overexpression of FOXQ1 alleviates hyperglycaemia and improves glucose tolerance in db/db diabetic and DIO mice
We next hypothesised that hepatic FOXQ1 deficiency contributes to the hyperglycaemic phenotype in diabetic mice and that FOXQ1 rescue would sufficiently restore glucose homeostasis. To test this hypothesis, we injected Ad-Foxq1 into db/db mice via the tail vein, which led to an increase in FOXQ1 expression levels in the liver (Fig. 3a), but did not affect FOXQ1 expression in the other tissues examined, including muscle and abdominal white adipose tissue (data not shown). Consistent with the results obtained in primary hepatocytes, the overexpression of FOXQ1 in mouse liver suppressed the expression of gluconeogenic genes, including Pgc-1α, G6pc and Pck1 (Fig. 3a). Subsequently, Ad-Foxq1 infection in the liver lowered fasting blood glucose and insulin levels compared with control (Ad-gfp treatment of db/db mice) (Fig. 3b,c). The glucose tolerance test (GTT) experiments indicated that hepatic overexpression of FOXQ1 caused a significant improvement in glucose excursion after glucose challenge (Fig. 3d). We also evaluated insulin tolerance and pyruvate tolerance in these mice. Notably, although blood glucose levels at each time point examined in these experiments in Ad-Foxq1-infected db/db mice were lower than that in control Ad-gfp -infected db/db mice, overexpression of FOXQ1 in db/db mice did not alter insulin sensitivity and pyruvate tolerance test (Fig. 3e,f), since baseline glucose levels in these mice were lower than those in control mice. In addition, FOXQ1 rescue in the livers of db/db mice did not significantly affect body weight, circulating or hepatic cholesterol, and NEFA concentration. However, FOXQ1 overexpression significantly reduced hepatic and serum triacylglycerol levels, and liver weight/body weight ratio (ESM Table 2). We also explored the molecular mechanism underlying the FOXQ1-mediated decrease in hepatic and serum triacylglycerol levels. We found that hepatic FOXQ1 overexpression markedly inhibited expression of lipogenic genes, including Srebp-1c (also known as Srebf1), Fas and Acc (ESM Fig. 3).
Adenovirus-mediated overexpression of FOXQ1 in db/db mice decreases blood glucose levels and improves glucose tolerance. db/db mice were injected with Ad-gfp (control, black bars/circles) or Ad-Foxq1 (white bars/squares). (a) qPCR analysis of mRNA levels and western blot analysis of protein levels of FOXQ1 and gluconeogenic genes/proteins in the liver of db/db mice 8 days after infection with Ad-gfp or Ad-Foxq1. (b) Plasma glucose levels in 6 h fasted db/db mice described in (a). (c) Plasma insulin concentrations in db/db mice as described in (a). (d) The blood glucose concentrations and AUC during the GTT in db/db mice infected with Ad-gfp and Ad-Foxq1. (e, f) Blood glucose concentrations during the (e) ITT and (f) pyruvate tolerance test in db/db mice infected with Ad-gfp and Ad-Foxq1. Data are presented as mean ± SEM. n = 6 for all. *p < 0.05, **p < 0.01 vs Ad-gfp treatment
Similar results were observed in DIO mice (ESM Fig. 4). Hepatic FOXQ1 rescue in DIO mice suppressed the expression of gluconeogenic genes, decreased blood glucose levels, improved glucose tolerance (Fig. 4a–c), and reduced hepatic triacylglycerol levels (ESM Table 3). However, FOXQ1 overexpression did not alter insulin sensitivity (Fig. 4d).
Adenovirus-mediated overexpression of FOXQ1 in DIO mice ameliorates hyperglycaemia and glucose intolerance. DIO mice were injected with Ad-gfp (control, black bars/circles) or Ad-Foxq1 (white bars/squares). (a) qPCR analysis of mRNA levels of Foxq1, Pgc-1α, G6pc and Pck1 in the livers of DIO mice 8 days after infection with Ad-gfp or Ad-Foxq1. (b) Plasma glucose levels in 6 h fasted DIO mice as described in (a). (c) Blood glucose concentrations and calculated AUC during the GTT in DIO mice infected with Ad-gfp and Ad-Foxq1. (d) Blood glucose concentrations during the ITT in DIO mice infected with Ad-gfp and Ad-Foxq1. Data are presented as mean ± SEM. n = 6 for all. *p < 0.05 vs Ad-gfp treatment
Together, these results suggest that hepatic FOXQ1 rescue in db/db diabetic and DIO mice inhibits hepatic gluconeogenesis, eventually resulting in reduced blood glucose levels and improved glucose intolerance.
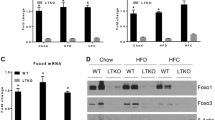
Hepatic silencing of FOXQ1 increases the gluconeogenic programme in wild-type C57BL/6 J mice
To further confirm the inhibitory effects of FOXQ1 on hepatic glucose production, we generated an adenovirus expressing Foxq1-specific shRNA (Ad-shFoxq1) and injected Ad-shFoxq1 into normal C57BL/6 J mice via the tail vein. The injection of Ad-shFoxq1 significantly reduced hepatic Foxq1 expression levels compared with the Ad-shCtrl treatment, whereas Ad-shFoxq1 treatment did not affect Foxq1 gene expression in white adipose tissue or skeletal muscle (data not shown). The hepatic expression levels of the gluconeogenic genes were increased in Ad-shFoxq1 treated mice compared with control (Ad-shCtrl) (Fig. 5a). The mice infected with Ad-shFoxq1 displayed higher fasting blood glucose (Fig. 5b). Moreover, hepatic FOXQ1 knockdown impaired glucose tolerance (Fig. 5c), while it did not alter insulin sensitivity (Fig. 5d). However, knockdown of FOXQ1 did not significantly affect body weight, liver weight/body weight ratio, circulating or hepatic cholesterol, NEFA and triacylglycerol concentration (ESM Table 4), although Srebp-1c was modestly increased in these mice (ESM Fig. 5). These data suggest that FOXQ1 exerts a significant regulatory effect on hepatic gluconeogenesis.
Adenovirus-mediated knockdown of FOXQ1 in C57BL/6J mice increases blood glucose levels and impairs glucose tolerance. Mice were injected with Ad-shCtrl (control, black bars/circles) or Ad-shFoxq1 (white bars/squares). (a) qPCR analysis of mRNA levels of Foxq1, Pgc-1α, G6pc and Pck1 and protein levels of FOXQ1, PGC-1α and G6Pase in the livers of C57BL/6J mice 8 days after infection. (b) Plasma glucose levels in C57BL/6 J mice 8 days after infection. (c) Blood glucose levels and AUC during the GTT in C57BL/6J mice on day 5 after infection. (d) Blood glucose levels during the insulin tolerance test (ITT) in mice on day 7 after infection. Data are presented as mean ± SEM. n = 7 for all. *p < 0.05, vs Ad-shCtrl treatment
FOXQ1 inhibits G6pc promoter activity through blocking FOXO1 binding to IRE via a physical interaction
FOXQ1 belongs to the forkhead family of transcription factors which have a conserved forkhead/winged helix DNA-binding domain (DBD) [30]. The optimal DNA-binding sequence for FOXQ1 has been identified [31]. FOXQ1 recognises the same consensus sequence (5-[A/T]TGTTTA[G/T]-3) as FOXO1. FOXO1 has been shown to activate expression of Pck1 and G6pc through directly binding to IREs mapped in the promoters of these target genes [14, 15]. However, FOXQ1 may act as a transcriptional repressor [18, 22]. Thus, we first hypothesised that FOXQ1 may compete with FOXO1 for binding to the IRE of gluconeogenic genes. To test this hypothesis, we first performed a promoter-luciferase reporter gene assay in HepG2 cells. The transfection of FOXO1 expression plasmid alone into HepG2 cells led to a marked activation of the G6pc-Luc-922 reporter gene, whereas overexpression of FOXQ1 alone did not significantly inhibit its activity (Fig. 6a). Next we performed EMSAs to determine whether FOXQ1 directly binds to the IRE of G6pc in vitro. As expected, FOXO1 proteins reacted with the biotin-labelled oligonucleotide probes, resulting in a shifted band. However, FOXQ1 proteins failed to bind to the DNA probes. Moreover, co-transfection of FOXQ1 reduced the formation of FOXO1–DNA protein complexes (Fig. 6a). Additionally, our chromatin immunoprecipitation (ChIP) analysis failed to detect FOXQ1 proteins binding to the IRE of G6pc in primary hepatocytes (data not shown). These results ruled out the possibility that FOXQ1 competes with FOXO1 for direct binding to the IRE of gluconeogenic genes.
FOXQ1 inhibits FOXO1-mediated stimulation of G6pc expression. Luciferase reporter gene assay was performed in HepG2 cells transfected with the indicated plasmids (white bars, pcDNA3.1; black bars, FOXO1 expression plasmid; diagonally striped bars, FOXQ1 expression plasmid; grey bars, FOXO1+ FOXQ1 expression plasmids). (a) Luciferase reporter gene assay and EMSA results shows that FOXQ1 blocks FOXO1 proteins binding to the IRE of the G6pc promoter. (b) Luciferase reporter gene assay shows that FOXQ1 completely blocks FOXO1-stimulated activity of pGL2–3×IREs in HepG2 cells. (c) Nucleotide sequence of wild-type and mutant IRE in the G6pc gene promoter and luciferase reporter gene assay; the reporter gene is driven by a series of truncated G6pc promoters. (d) Quantitative PCR assay of G6pc gene expression in HepG2 cells transfected with FOXO1 and/or FOXQ1 expression plasmids and control (pcDNA3.1) plasmids. (e) Western blot of FOXO1 and Myc and quantitative PCR of G6pc in primary mouse hepatocytes infected with indicated adenoviruses (Ad-Foxq1 expresses myc-tagged Foxq1). (f) Glucose production of primary mouse hepatocytes infected with indicated adenoviruses. (g) ChIP assay using primary mouse hepatocytes infected with adenoviruses (Ad-gfp, white bars; Ad-Foxo1, black bars; Ad-Foxo1+Ad-Foxq1, grey bars), demonstrating that FOXQ1 inhibits FOXO1 binding to IRE of the G6pc gene promoter. Results are presented as means ± SEM of 3 replication experiments. *p < 0.05, **p < 0.01. Relative luciferase activity (RLA) was corrected for Renilla luciferase activity and normalised to the control activity
Seoane et al reported that FOXG1 abolishes FOXO-mediated induction of cyclin-dependent kinase inhibitor (p21Cip1) through antagonising FOXO1 activity [32]. This prompted us to explore the possibility that FOXQ1 may directly interact with FOXO1 in vivo, thereby forming a complex and interfering with FOXO1 activity. To test this idea we first examined transactivation of FOXO1 and FOXQ1 using a luciferase reporter gene driven by three tandem repeats of IRE (pGL2–3×IRE) in HepG2 cells. The transfection of FOXO1 into HepG2 cells caused an approximately sevenfold activation of the pGL2–3×IREs reporter gene (Fig. 6b). Co-transfection of FOXQ1 expression plasmids completely abolished FOXO1 activation of pGL2–3×IREs reporter (Fig. 6b).
Similar results were obtained using G6pc-Luc-922 reporter (Fig. 6c). To further confirm that IRE is essential for FOXO1- and FOXQ1-mediated G6pc gene transcription, a series of truncated segments of the G6pc promoter were fused to the luciferase gene (G6pc-Luc-300, G6pc-Luc-133) and transfected into HepG2 cells. As a result, FOXQ1 still antagonised FOXO1 action on the G6pc-Luc-300 reporter, which contained the IRE. However, when the reporter gene was further truncated to −133 bp, deleting the IRE, FOXO1 and FOXQ1 failed to control its activity (Fig. 6c). Similar results were obtained for G6pc-Luc-922 mutant (mut) reporter (Fig. 6c), in which the IRE of the G6pc promoter was mutated.
To further confirm the interplay between FOXO1 and FOXQ1 in cells, HepG2 cells were transfected with FOXO1 and/or FOXQ1 expression plasmids. As expected, overexpression of FOXO1 stimulated G6pc expression, while FOXQ1 inhibited FOXO1-induced G6pc gene expression (Fig. 6d). Similar data were observed in primary hepatocytes infected with Ad-Foxo1 and/or Ad-Foxq1 (Fig. 6e). Functional studies also confirmed that FOXQ1 antagonised FOXO1 stimulation of cellular glucose production in primary hepatocytes (Fig. 6f).
Finally, we performed a ChIP assay to examine whether FOXQ1 proteins block FOXO1 binding to the IRE of G6pc. As a result, Ad-Foxo1 infection promoted FOXO1 binding to the IRE of G6pc, while co-infection by Ad-Foxq1 completely blocked FOXO1 binding (Fig. 6g). Collectively, these results suggest that FOXQ1 antagonises FOXO1-mediated transcription of gluconeogenic genes.
To further characterise how FOXQ1 antagonises FOXO1 activity, we sought to investigate whether FOXQ1 directly binds to FOXO1. To test this possibility, we transiently expressed myc-HA-tagged FOXO1 and myc-tagged FOXQ1 in HEK293A cells. Immunoprecipitation with anti-FOXQ1 antibodies also pulled down FOXO1 protein, as assessed by western blot analysis (Fig. 7a), indicating that these two proteins formed a complex in vivo. Given that FOXO3, a member of the FOXO subfamily, shares high homology with FOXO1, we next explored whether FOXQ1 also binds to FOXO3 in vivo. However, as shown in Fig. 7b, we could not detect the interaction between flag-tagged FOXO3 and FOXQ1, indicating that FOXQ1 specifically interacts with FOXO1 in vivo.
FOXQ1 proteins directly bind the DBD of FOXO1. (a) Co-immunoprecipitation (CoIP) assay showing that FOXQ1 interacts with FOXO1, (b) but not with FOXO3, in vivo. HEK293A cells were transfected with plasmid (a) encoding myc-tagged FOXQ1 and myc-HA-tagged FOXO1 or (b) flag-tagged FOXO3 plasmids and immunoprecipitates with anit-FOXQ1 were immunoblotted with the corresponding antibodies. (c) GST pull down assay showing FOXQ1 physically interacts with FOXO1 in vitro. (d) Schematic representation of the primary structure of FOXO1 proteins and GST pull down assay showing that the DBD of FOXO1 interacts with FOXQ1. NLS, Nuclear localisation signal
To further confirm a direct physical interaction between FOXQ1 and FOXO1, we performed an in vitro interaction assay using recombinant GST-FOXQ1 fusion proteins. This analysis showed that GST-FOXQ1, but not GST alone, bound to full-length FOXO1 (Fig. 7c). To identify the domain of FOXO1 responsible for the interaction with FOXQ1, three flag-tagged fusion plasmids were generated containing amino acids 1–145 (N-terminus), amino acids 146–274 (DBD) and amino acids 1–274 (N+DBD) of FOXQ1. Subsequent GST pull down experiments indicated that the DBD of FOXO1 mediates its interaction with FOXQ1 (Fig. 7d). These data illustrate that FOXQ1 directly interacts with the DBD of FOXO1 and inhibits FOXO1-mediated induction of G6pc expression.
Based on our data, combined with that of previous reports [33, 34], we propose a model for the mechanism of FOXQ1 action on hepatic gluconeogenesis (Fig. 8).
Proposed model of FOXQ1-mediated repression of hepatic gluconeogenesis. Under feeding conditions, activated Akt (by insulin) phosphorylates FOXO1, which induces 14-3-3 protein binding to phospho-FOXO1 and promotes its nuclear export, subsequently leading to its proteasomal degradation in the cytoplasm. Meanwhile, FOXQ1 proteins are increased and form complexes with non-phospho-FOXO1, blocking FOXO1 activity on hepatic gluconeogenesis and maintaining glucose homeostasis. However, under certain pathophysiological conditions (insulin resistance and diabetes) or the fasting state, Akt activity is inhibited and FOXQ1 protein levels decrease, allowing FOXO1 to bind to gluconeogenic genes, contributing to hepatic glucose production
Discussion
Members of the forkhead box family share a highly conserved DBD, and a core FOXQ1 binding sequence (TGTTTA) has been identified [31], which is highly similar to the IRE identified in the G6pc gene promoter. However, our data (from luciferase reporter gene assay, EMSA, ChIP and GST pull down experiments) indicate that FOXQ1 exerts its inhibitory effects on the expression of gluconeogenic genes through blocking FOXO1 binding to these genes via a physical interaction with the DBD of FOXO1, rather than by directly interacting with the promoter region of gluconeogenic genes. Notably, although our data show that FOXQ1 antagonises FOXO1 activity on gluconeogenic genes, we cannot rule out the possibility that FOXQ1 may regulate expression of gluconeogenic genes via other mechanisms. For example, FOXQ1 may control expression and/or activity of other factors, including hepatic nuclear factor-4 (HNF4), the glucocorticoid receptor (GR) and cAMP responsive element binding protein (CREB), which have been shown to regulate gluconeogenic genes [3, 35]. Further studies are needed to clarify this.
We found that hepatic FOXQ1 is regulated by fasting and pathophysiological status (insulin resistance and diabetes). In the fasted state insulin levels drop while secretion of glucagon and glucocorticoids increase, resulting in increased hepatic gluconeogenesis [3, 36]. Our data indicates that glucocorticoids, rather than glucagon or insulin, regulate the expression of FOXQ1. Moreover, glucocorticoid levels have been shown to be increased in insulin-resistant and glucose-intolerant individuals [37, 38]. Notably, further studies are required to determine whether FOXQ1 is required for the effects of glucocorticoids on gluconeogenic gene expression in primary hepatocytes after knocking down FOXQ1 expression. Thus, decreased FOXQ1 expression in the liver of mice with insulin resistance might result from increased glucocorticoid levels. Similarly, the reduction in FOXQ1 expression in fasted normal mice may result from an increase in glucocorticoid effect, rather than loss of insulin effect. Glucocorticoids mediate their physiological effects through binding to the GR. Upon glucocorticoid binding in the cytosol, GR translocates into the nucleus where it regulates expression of its target genes via direct DNA binding [39]. Further studies are required to determine whether glucocorticoids/GR inhibit FOXQ1 expression in a direct or indirect manner.
The role of FOXO1 in regulating hepatic lipid homeostasis and metabolism remains controversial. Previous studies indicate that FOXO1 overexpression in the liver increased hepatic triacylglycerol content, owing to increased triacylglycerol synthesis [40, 41], while recent studies suggest that FOXO proteins suppress hepatic lipogenesis and triacylglycerol levels [42, 43]. In this study we show that overexpression of FOXQ1 decreases hepatic and serum triacylglycerol contents in db/db and DIO mice. Further studies are needed to clarify the molecular mechanisms underlying FOXQ1-mediated changes in hepatic or serum triacylglycerol levels.
Decreased FOXQ1 expression might be an underlying factor for increased hepatic gluconeogenesis and the development of diabetes. Thus our study provides a novel therapeutic target for the treatment of type 2 diabetes, demonstrating that small molecules that induce increases in FOXQ1 levels may have potential for use as treatment of this disease.
Abbreviations
- Ad-gfp :
-
Adenovirus containing green fluorescent protein
- Ad-Foxq1 :
-
Adenovirus expressing FOXQ1
- Ad-shCtrl:
-
Control adenovirus expressing short-hairpin RNA against luciferase
- Ad-shFoxq1 :
-
Adenovirus expressing short-hairpin RNA against FOXQ1
- ChIP:
-
Chromatin immunoprecipitation
- DBD:
-
DNA-binding domain
- DEX:
-
Dexamethasone
- DIO:
-
Diet-induced obese
- EMSA:
-
Electromobility shift assay
- FOX:
-
Forkhead box
- FSK:
-
Forskolin
- G6pase:
-
Glucose-6-phosphatase, catalytic
- GR:
-
Glucocorticoid receptor
- GST:
-
Glutathione S-transferase
- GTT:
-
Glucose tolerance test
- IRE:
-
Insulin response element
- ITT:
-
Insulin tolerance test
- PGC-1α:
-
Peroxisome proliferator-activated receptor, gamma, co-activator 1 alpha
- qPCR:
-
Quantitative PCR
- shRNA:
-
Short-hairpin RNA
References
Saltiel AR, Kahn CR (2001) Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414:799–806
Puigserver P, Rhee J, Donovan J et al (2003) Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 423:550–555
Yoon JC, Puigserver P, Chen G et al (2001) Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413:131–138
Pilkis SJ, Granner DK (1992) Molecular physiology of the regulation of hepatic gluconeogenesis and glycolysis. Annu Rev Physiol 54:885–909
Brunet A, Bonni A, Zigmond MJ et al (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857–868
Dentin R, Liu Y, Koo SH et al (2007) Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449:366–369
Dong XC, Copps KD, Guo S et al (2008) Inactivation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metab 8:65–76
Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D (2007) Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab 6:208–216
Nakae J, Biggs WH 3rd, Kitamura T et al (2002) Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet 32:245–253
Li X, Monks B, Ge Q, Birnbaum MJ (2007) Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature 447:1012–1016
Accili D, Arden KC (2004) FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell 117:421–426
Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM (1999) Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398:630–634
Biggs WH 3rd, Meisenhelder J, Hunter T, Cavenee WK, Arden KC (1999) Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci U S A 96:7421–7426
Onuma H, Vander Kooi BT, Boustead JN, Oeser JK, O’Brien RM (2006) Correlation between FOXO1a (FKHR) and FOXO3a (FKHRL1) binding and the inhibition of basal glucose-6-phosphatase catalytic subunit gene transcription by insulin. Mol Endocrinol 20:2831–2847
O’Brien RM, Noisin EL, Suwanichkul A et al (1995) Hepatic nuclear factor 3- and hormone-regulated expression of the phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein 1 genes. Mol Cell Biol 15:1747–1758
Frank S, Zoll B (1998) Mouse HNF-3/fork head homolog-1-like gene: structure, chromosomal location, and expression in adult and embryonic kidney. DNA Cell Biol 17:679–688
Hong HK, Noveroske JK, Headon DJ et al (2001) The winged helix/forkhead transcription factor Foxq1 regulates differentiation of hair in satin mice. Genesis 29:163–171
Hoggatt AM, Kriegel AM, Smith AF, Herring BP (2000) Hepatocyte nuclear factor-3 homologue 1 (HFH-1) represses transcription of smooth muscle-specific genes. J Biol Chem 275:31162–31170
Verzi MP, Khan AH, Ito S, Shivdasani RA (2008) Transcription factor foxq1 controls mucin gene expression and granule content in mouse stomach surface mucous cells. Gastroenterology 135:591–600
Kaneda H, Arao T, Tanaka K et al (2010) FOXQ1 is overexpressed in colorectal cancer and enhances tumorigenicity and tumor growth. Cancer Res 70:2053–2063
Qiao Y, Jiang X, Lee ST, Karuturi RK, Hooi SC, Yu Q (2011) FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res 71:3076–3086
Zhang H, Meng F, Liu G et al (2011) Forkhead transcription factor foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Cancer Res 71:1292–1301
Feng J, Zhang X, Zhu H, Wang X, Ni S, Huang J (2012) FoxQ1 overexpression influences poor prognosis in non-small cell lung cancer, associates with the phenomenon of EMT. PLoS One 7:e39937
Luo J, Deng ZL, Luo X et al (2007) A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc 2:1236–1247
Zhang H, Chen Q, Yang M et al (2013) Mouse KLF11 regulates hepatic lipid metabolism. J Hepatol 58:763–770
Kong X, Wang R, Xue Y et al (2010) Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One 5:e11707
Wang R, Kong X, Cui A et al (2010) Sterol-regulatory-element-binding protein 1c mediates the effect of insulin on the expression of Cidea in mouse hepatocytes. Biochem J 430:245–254
Le Lay J, Kaestner KH (2010) The Fox genes in the liver: from organogenesis to functional integration. Physiol Rev 90:1–22
Cederberg A, Gronning LM, Ahren B, Tasken K, Carlsson P, Enerback S (2001) FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell 106:563–573
Bieller A, Pasche B, Frank S et al (2001) Isolation and characterization of the human forkhead gene FOXQ1. DNA Cell Biol 20:555–561
Overdier DG, Porcella A, Costa RH (1994) The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol Cell Biol 14:2755–2766
Seoane J, Le HV, Shen L, Anderson SA, Massague J (2004) Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117:211–223
Tzivion G, Dobson M, Ramakrishnan G (2011) FoxO transcription factors; regulation by AKT and 14-3-3 proteins. Biochim Biophys Acta 1813:1938–1945
Cheng Z, White MF (2011) Targeting Forkhead box O1 from the concept to metabolic diseases: lessons from mouse models. Antioxid Redox Signal 14:649–661
Herzig S, Long F, Jhala US et al (2001) CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413:179–183
Quinn PG, Yeagley D (2005) Insulin regulation of PEPCK gene expression: a model for rapid and reversible modulation. Curr Drug Targets Immune Endocr Metabol Disord 5:423–437
Phillips DI, Barker DJ, Fall CH et al (1998) Elevated plasma cortisol concentrations: a link between low birth weight and the insulin resistance syndrome? J Clin Endocrinol Metab 83:757–760
Reynolds RM, Walker BR, Syddall HE, Whorwood CB, Wood PJ, Phillips DI (2001) Elevated plasma cortisol in glucose-intolerant men: differences in responses to glucose and habituation to venepuncture. J Clin Endocrinol Metab 86:1149–1153
Lemke U, Krones-Herzig A, Berriel Diaz M et al (2008) The glucocorticoid receptor controls hepatic dyslipidemia through Hes1. Cell Metab 8:212–223
Matsumoto M, Han S, Kitamura T, Accili D (2006) Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J Clin Invest 116:2464–2472
Qu S, Altomonte J, Perdomo G et al (2006) Aberrant forkhead box O1 function is associated with impaired hepatic metabolism. Endocrinology 147:5641–5652
Haeusler RA, Hartil K, Vaitheesvaran B et al (2014) Integrated control of hepatic lipogenesis versus glucose production requires FoxO transcription factors. Nat Commun 5:5190
Zhang K, Li L, Qi Y et al (2012) Hepatic suppression of Foxo1 and Foxo3 causes hypoglycemia and hyperlipidemia in mice. Endocrinology 153:631–646
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported by the Major State Basic Research Development Program of China (973 programme grant 2012CB517504), the National Natural Science Foundation of China (Grant no. 81471049 and no. 81170763) and the Natural Science Foundation of Beijing Municipality (Grant no. 5142018).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
YCh contributed to the conception and design of the study. YCu, AQ, TJ, YX, HZ, YZ and AC designed and performed studies and analysed data. FF contributed to analysis of data. YCu and YCh wrote the manuscript. All authors critically revised the manuscript and approved the final version. YCu and YCh are the guarantors of this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM
(PDF 512 kb)
Rights and permissions
About this article
Cite this article
Cui, Y., Qiao, A., Jiao, T. et al. The hepatic FOXQ1 transcription factor regulates glucose metabolism in mice. Diabetologia 59, 2229–2239 (2016). https://doi.org/10.1007/s00125-016-4043-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-016-4043-z