Abstract
Aims/hypothesis
Pancreatic beta cells play a central role in the control of glucose homeostasis by secreting insulin to stimulate glucose uptake by peripheral tissues. Understanding the molecular mechanisms that control beta cell function and plasticity has critical implications for the pathophysiology and therapy of major forms of diabetes. Selective gene inactivation in pancreatic beta cells, using the Cre-lox system, is a powerful approach to assess the role of particular genes in beta cells and their impact on whole body glucose homeostasis. Several Cre recombinase (Cre) deleter mice have been established to allow inactivation of genes in beta cells, but many show non-specific recombination in other cell types, often in the brain.
Methods
We describe the generation of Ins1 Cre and Ins1 CreERT2 mice in which the Cre or Cre-oestrogen receptor fusion protein (CreERT2) recombinases have been introduced at the initiation codon of the Ins1 gene.
Results
We show that Ins1 Cre mice induce efficient and selective recombination of floxed genes in beta cells from the time of birth, with no recombination in the central nervous system. These mice have normal body weight and glucose homeostasis. Furthermore, we show that tamoxifen treatment of adult Ins1 CreERT2 mice crossed with Rosa26-tdTomato mice induces efficient recombination in beta cells.
Conclusions/interpretation
These two strains of deleter mice are useful new resources to investigate the molecular physiology of pancreatic beta cells.
Similar content being viewed by others
Introduction
Pancreatic beta cells participate in glucose homeostasis by secreting insulin in response to rises in glycaemia to stimulate glucose uptake by peripheral tissues. To maintain this capacity over a lifetime, the insulin secretion capacity of individual beta cells as well as their number may be modulated to compensate for the development of insulin resistance in target tissues. Failure of this beta cell adaptive capacity is a major cause of the development of diabetic hyperglycaemia. On the other hand, the enhancement of natural mechanisms of beta cell adaptation might prove useful for regenerative therapies in autoimmune type 1 diabetes. Understanding the molecular basis of beta cell plasticity is therefore a central focus in diabetes research [1].
Genetically modified mice are being used extensively to study the role of specific genes in diabetes pathophysiology [2–5], and several genetic systems have been developed to overexpress or inactivate genes selectively in beta cells [6–12]. In particular, recent recombinant technologies have provided the possibility to selectively inactivate genes of interest in a tissue- and time-dependent manner, as critically reviewed recently [13]. A particularly useful approach consists in flanking the DNA sequence to be deleted by tandemly oriented 34 nucleotide-long lox sequences, which are recognised by Cre recombinase (Cre), a recombination enzyme that leads to the elimination of the sequence present between the two lox sites. Modification of the target DNA sequence with lox sites is performed by homologous recombination in embryonic stem cells, and the mice that are ultimately generated are crossed with mice that express Cre under a tissue-specific promoter to allow tissue-specific gene deletion. The use of Cre-oestrogen receptor fusion protein (CreERT2) [14], which is Cre fused to a modified form of the ligand-binding domain of the human oestrogen receptor that can be activated by tamoxifen, allows precise time-controlled genetic recombination.
One key aspect of the Cre-lox system is that transgenic expression of the Cre gene must be specific to the desired target tissue. Deletion in beta cells has been performed using mice expressing Cre under the control of various promoters such as that of the rat Ins2 gene [12, 15], the mouse Ins1 promoter [16] or the human insulin promoter [17]. In contrast to the mouse Ins2 gene, which is also expressed in the embryonic and adult brain, the Ins1 gene is selectively expressed by the beta cells [18, 19], thereby providing a potential specificity advantage to drive Cre reporter expression only in beta cells. Existing transgenics that use insulin gene regulatory regions typically allow for beta cell gene inactivation; however, they often also lead to expression in brain regions, mostly the hypothalamus for the Ins2 promoter, leading to phenotypes that are complicated by the non-specific site of expression of the recombinase. Other approaches have used the promoters of transcription factor genes expressed at different stages of beta cell development, such as Pdx-1, Neurog3, Nkx2.2, Pax4, Pax6 or Isl-1 (for detailed review of these lines, see [13]). Although all of these lead to Cre expression in beta cells, they also lead to gene deletion either in islet non-beta cells or diverse non-pancreatic cellular lineages, including widely used Pdx1-Cre mouse lines that induce recombination in the exocrine pancreas. There is thus a need for more selective Cre lines for gene inactivation in beta cells.
The design of a mouse model that transcribes Cre exclusively in beta cells faces several obstacles. One is that gene regulatory sequences are not exclusively encoded in the 5′ regions of genes but are often determined or modulated by long-range cis-regulatory interactions, most of which have not been annotated. This obstacle is best addressed by inserting the recombinase in the endogenous genomic locus of interest, rather than using predefined promoter fragments. Another complexity is that most genes that are enriched in beta cells are also expressed in other cell types, some of which are central to glucose homeostasis. Notably, of the two non-allelic mouse insulin genes, Ins2 is the most abundantly expressed in beta cells, yet it is also expressed in hypothalamus and brain [18]. Thus, even genetic models that succeed in recapitulating Ins2 expression will unavoidably lead to expression outside the beta cells, while recapitulation of Ins1 expression is expected to lead to selective beta cell expression
Here, we generated mice in which Cre was introduced at the initiator codon of the Ins1 locus by homologous recombination technology so as to ensure faithful expression of the recombinase in beta cells. We show that this approach allowed for highly efficient expression of Cre or CreERT2 recombinase in beta cells and that the specificity of expression was guaranteed with no expression in hypothalamus or other brain areas and with recombination as early as the time of birth.
Methods
Mice Ins1 Cre (Ins1 tm1(cre)Thor) and Ins1 CreERT2 (Ins1 tm1(CreERT2)Thor) knock-in mice were generated by Genoway (Lyon, France) and kept on a C57Bl/6J genetic background. Briefly, a targeting vector was created by inserting the Cre or CreERT2 recombinase genes by homologous recombination in the second exon of the Ins1 gene so that the coding region of the recombinase starts at the initiation codon and replaces the Ins1 coding sequence (Fig. 1). Following transfection of the targeting vector into embryonic stem cells, negative and positive selections were performed by diphtheria toxin and neomycin (neo) selection, respectively. Cells with confirmed homologous recombination were used to generate chimeric mice. These were crossed with C57Bl/6J mice, and mice with germline transmission were crossed with C57Bl/6J flippase (Flp) deleter mice to eliminate the neo cassette. Mice with germline transmission of the recombined allele and deletion of the neo cassette were then crossed with Rosa26-eYFP mice [20] or Rosa26-tdTomato mice [21]. Animals were housed four to five per cage at 23°C on a 12 h light/dark cycle and were fed a standard rodent chow (Diet 3436; Provimi Kliba, Penthallaz, Switzerland). To induce recombination in Ins1 CreERT2 crossed with Rosa26 reporter mice, tamoxifen (T5648; Sigma-Aldrich, St Louis, MO, USA) was dissolved in corn oil at 20 mg/ml, and 2 mg was injected subcutaneously four times over a 2 week period.
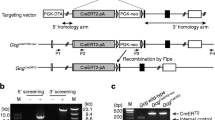
Generation of Ins1Cre and Ins1 CreERT2 mice. (a) Structure of the Ins1 locus, of the targeting vector, of the targeted allele, and of the Ins1 Cre locus following flipase (Flp)-dependent removal of the flipase recognition site (Frt)-flanked neo cassette. DTA, diphtheria toxin receptor gene; Ex, exon; ATG, translation initiation codon. (b) The Ins1 CreERT2 allele was obtained using the targeting strategy depicted in (a). (c) Structure of the Rosa26-eYFP/tdTomato transgene and localisation of the primers (arrows) used for PCR analysis of the recombined locus. CMV/A, cytomegalovirus/actin promoter; (d) PCR analysis shows recombination in the pancreas but not in the liver, cortex, hypothalamus or cerebellum of Ins1 Cre/+ ;Rosa26-eYFP. No recombination was observed in the pancreas of Rosa26-eYFP littermates; amplification of the Gapdh gene is used as a control
Genotyping
Mouse genotyping was performed by PCR analysis using primers listed in electronic supplementary material (ESM) Table 1.
Histological and immunodetection procedures
For immunodetection of enhanced yellow fluorescent protein (eYFP), tdTomato, insulin, glucagon, pancreatic polypeptide and somatostatin in the pancreas or in the brain, mice were fixed by perfusion with cold 4% paraformaldehyde in sodium phosphate buffer (0.1 mol/l, pH 7.4). The organs were dissected and kept for 4 h in the same paraformaldehyde solution before overnight incubation in 30% sucrose, then frozen in isopentane and stored at −80°C until used.
For immunofluorescence detection studies in eYFP reporter mice 20 μm cryosections were preincubated for 30 min in a permeabilisation blocking buffer (0.1 mol/l PBS, pH 7.4, 3% BSA A8022, 0.3% Triton X-100, 9002-93-1; Sigma-Aldrich), then incubated overnight at 4°C with a rabbit polyclonal antibody to green fluorescent protein (GFP) (ab290, diluted 1:2,000; Abcam, Cambridge, UK), a polyclonal guinea pig anti-insulin (AO564, diluted 1:400; Dako, Baar, Switzerland) or anti-glucagon (4031-01F, diluted 1:500; Linco Research, St Charles, MO, USA), a polyclonal anti-pancreatic polypeptide (diluted 1:1,000; Linco Research) or a polyclonal rabbit anti-somatostatin (A0566, diluted 1:200; Dako). After washing, the sections were then incubated for 90 min with Alexa Fluor 488 goat anti-guinea pig IgG (H+L) (A11073, diluted 1:200; Invitrogen, LuBioScience, Lucerne, Switzerland) or with a goat anti-rabbit immunoglobulin-Cy3 conjugate (111-165-144, diluted 1/100; Jackson ImmunoResearch, West Grove, PA, USA), or for the Rosa26-tdTomato mice with Alexa Fluor 488 goat anti-rabbit IgG (H+L) (A-11008, diluted 1:200; Invitrogen). Immunofluorescence analysis of tdTomato was carried out in 3 μm paraffin sections, essentially as described [22, 23], using a rabbit polyclonal anti-red fluorescent protein (600-401-379; Rockland Immunochemical, Gilbertsville, PA, USA).
For enzymatic detection of eYFP, after overnight incubation with primary antibody, sections were washed and incubated for 90 min with a biotinylated goat anti-rabbit antibody (BA-1000, diluted 1:750; Vector Laboratories, Burlingame, CA, USA), incubated for 30 min with a biotin–avidin complex (Vector) and stained using 3,3′-diaminobenzidine tetrahydrochloride (Sigma-Aldrich).
Sections were observed using an Axio Imager D1 microscope (Carl Zeiss, Oberkochen, Germany) interfaced with Axiovision software (Carl Zeiss) or a 510 Meta inverted confocal laser scanning microscope (LSM) with LSM software (release 3.5; Zeiss).
Glucose tolerance tests
For glucose tolerance tests, 16 h fasted male and female mice of indicated ages were injected i.p. with glucose (2 g/kg), and blood glucose levels were determined from tail–tip bleedings at the indicated times relative to glucose injections. Blood glucose levels were measured using a glucometer (Ascensia Breeze 2; Bayer HealthCare, Switzerland).
Statistical analysis
Data are expressed as means±SD or SEM. Statistical analysis was performed using an unpaired two-tailed Student’s t test. Values of p < 0.05 were considered significant.
Study approval
All breeding and cohort maintenance performed in our animal facility and all experiments were approved by the Service Vétérinaire du Canton de Vaud and the Comitè Ètic d’Experimentació Animal of the University of Barcelona.
Results
Mouse generation
Generation of the Ins1 Cre and Ins1 CreERT2 mice followed the strategy described in Fig. 1a–c and the Methods section, allowing expression of Cre or CreERT2 from the translation initiation codon of the Ins1 gene. Embryonic stem cells with homologous recombination were used to generate chimeric mice. These were crossed with Flp deleter mice to remove the neo cassette. The resulting Ins1 Cre mice were crossed with Rosa26-eYFP mice to determine the anatomical sites of Cre-dependent lox site recombination. PCR analysis of the recombined Rosa26-eYFP locus (Fig. 1c) indicated recombination in whole pancreas DNA preparations but not in DNA extracted from liver, hypothalamus, cortex or cerebellum (Fig. 1d).
Assessment of the cellular sites of Cre-induced eYFP expression was then performed by histological analysis. In the pancreas, eYFP expression was only observed in islets as revealed by the endogenous fluorescence of eYFP or by immunofluorescence detection using anti-GFP antibodies (Fig. 2a, b). No eYFP expression was observed in the liver of the same mice (Fig. 2c, d), nor in the pancreas of control Rosa26-eYFP mice (Fig. 2e, f). Co-staining of the pancreas of Ins1 Cre/+ ;Rosa26-eYFP mice with islet hormones (insulin, glucagon, somatostatin or pancreatic polypeptide) showed that eYFP expression was specific to beta cells within pancreatic islets (Fig. 3a–d). We quantified the recombination efficiency in pancreatic cell subtypes using Ins1 Cre/+ ;Rosa26-tdTomato mice and found that 97.8 ± 1.2% of insulin-expressing cells showed expression of tdTomato, in contrast with 3.0 ± 0.3% of glucagon-expressing cells, 0.0% of somatostatin-expressing cells and <0.01% of CPA-1-expressing pancreatic acinar cells.
Expression of eYFP in pancreatic islets of Ins1 Cre/+ ;Rosa26-eYFP mice. (a, c, e) eYFP fluorescence is detected in the pancreatic islets (a) but not in the liver (c) of 12-week-old Ins1 Cre/+ ;Rosa26-eYFP mice; it is not detected in the pancreas of Rosa26-eYFP mice (e). (b, d, f) Indirect immunofluorescence detection of eYFP (eYFP-IF, red) on tissue sections corresponding to a, c and e, respectively. Scale bars, 50 μm (a–f)
To determine whether recombination could already be seen at the time of birth, pancreas sections were prepared from newborn Ins1 Cre/+ ;Rosa26-tdTomato mice and co-stained for insulin and glucagon. Figure 4a, b shows that expression of tdTomato was already present in beta cells at birth and no expression was detected in glucagon-producing alpha cells.
Because Cre mice previously generated to target the pancreatic beta cells often showed expression of the transgene in the brain, in particular in the hypothalamus, we verified expression of eYFP in different parts of the brain. PCR analysis (Fig. 1d) showed no detectable recombination of the Rosa26 locus in Ins1 Cre/+ ;Rosa26-eYFP mice in the hypothalamus, cortex or cerebellum. No eYFP fluorescence could be detected in the hypothalamus of Ins1 Cre/+ ;Rosa26-eYFP mice, as shown at two different bregma levels (ESM Fig. 1a, b); sections of the hypothalamus of control Rosa26-eYFP mice are shown in ESM Fig. 1c, d. As an alternative means of detecting recombination in the brain, we evaluated expression of eYFP by immunohistochemistry. No signal could be detected in any hypothalamic areas of Ins1 Cre/+ ;Rosa26-eYFP mice, as shown in Fig. 5a–d. Immunohistochemical detection of eYFP in the beta cells from Ins1 Cre/+ ;Rosa26-eYFP mice is shown as a positive control (Fig. 5e). Similarly, no positive signals could be detected in the cortex or thalamus of Ins1 Cre/+ ;Rosa26-eYFP mice (ESM Fig. 2a–d). Thus, these data are in agreement with the PCR data of Fig. 1c that show absence of recombination event in the brain of adult Ins1 Cre/+ ;Rosa26-eYFP mice.
No expression of eYFP in the hypothalamus of Ins1 Cre/+ ;Rosa26-eYFP mice as assessed by immunohistochemistry. Immunohistochemical detection of eYFP in the hypothalamus of Ins1 Cre/+ ;Rosa26-eYFP and control Rosa26-eYFP mice at bregma −0.82 (a, c) and −1.70 (b, d). (e) eYFP immunohistochemical detection in the pancreas from Ins1 Cre/+ ;Rosa26-eYFP mice. Scale bars, 100 μm. Arc, arcuate hypothalamic nucleus; LH, lateral hypothalamic area; 3 V, third ventricle
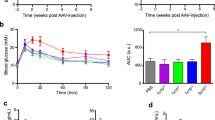
To determine whether introduction of the Cre gene in the Ins1 locus would cause any deregulation of glucose homeostasis or impact mouse growth, we generated cohorts of male and female C57Bl/6J heterozygous Ins1 Cre and littermate controls and followed them for 24 weeks. No difference in body weight gain could be observed between the control and Ins1 Cre mice of either sex (Fig. 6a). Random fed glycaemia levels were also identical (Fig. 6b), as was glucose tolerance assessed in heterozygous 12 week old male and female mice (Fig. 6c, d).
Normal body weight gain, glycaemic levels and glucose tolerance in heterozygous Ins1 Cre mice. (a) Body weight of male (white squares) and female (white triangles) Ins1 Cre mice and their control littermates (males, black squares; females, black triangles). (b) Fed glycaemic levels in 12-week-old male and female Ins1 Cre mice and their control littermates. (c, d) Glucose tolerance tests in 12-week-old male (c) and female (d) Ins1 Cre mice and their control littermates (white symbols, Ins1 Cre mice; black symbols, control littermates). White bars, Ins1 Cre mice; black bars, control littermates. Data are mean±SEM; n = 9–13
Finally, to determine whether recombination could be induced in the beta cells of adult Ins1 CreERT2/+ ;Rosa26-tdTomato mice, we tested several tamoxifen injection protocols. The data of Fig. 7 show that four injections of tamoxifen (2 mg in corn oil) over a 2 week period in 10 week old mice induced recombination in approximately 60–70% of beta cells. No expression of tdTomato could be observed in islets from corn oil-treated Ins1 CreERT2/+ ;Rosa26-tdTomato mice.
Tamoxifen-induced expression of tdTomato in beta cells of Ins1 CreERT2/+ ;Rosa26-tdTomato mice. (a–c) Ten-week-old Ins1 CreERT2/+ ;Rosa26-tdTomato mice were injected with 2 mg tamoxifen four times over a 2 week period. Expression of insulin (green) (a) and tdTomato (red) (b) was then assessed; (c) merged images. (d–f) Same as (a–c) except that mice were injected with vehicle (corn oil). Scale bars, 50 μm
Discussion
Here we describe the generation of two new mouse models for efficient and selective induction of lox-dependent recombination in pancreatic beta cells. The Ins1 Cre mice induce complete recombination by the time of birth in all beta cells, whereas the Ins1 CreERT2 mice can be used for tamoxifen-dependent gene recombination in adult mice.
Several Cre transgenic mice have already been established for beta cell-specific gene recombination. The present models use a knock-in strategy to express Cre from the endogenous Ins1 locus. This locus is known to have a more restricted beta cell expression than the Ins2 locus, which displays transient expression in the hypothalamus and other brain structures [24]. The use of the Ins2 promoter to drive Cre expression led to mouse models with compound genetic recombination in the islets and brain, leading to complicated interpretation of the phenotype of mice used [16]. Ins2 knock-in mouse lines have also been generated [25], but are similarly expected to recapitulate endogenous expression and result in ectopic expression in brain. On the other hand, two recent studies have reported transgenic models expressing Cre or CreER driven by Ins1 promoter sequences or by an Ins1 locus bacterial artificial chromosome [16, 26]. In keeping with our model, these mice lack Cre expression in the brain [16, 26]. Our model provides further advantages in that Cre is inserted in the native Ins1 locus and is thus theoretically controlled by all known and unknown native cis-regulatory sequences, providing greater ability to recapitulate the endogenous Ins1 expression pattern. We have indeed verified that recombination is efficiently induced in pancreatic islet beta cells at birth, whereas no recombination could be observed in the brain. Furthermore, we report an inducible Ins1 CreERT2 mouse line that is capable of recombination in 60–70% of the beta cells. Importantly, there was no leakiness of CreERT2 expression as judged by the absence of tdTomato expression in non-treated or vehicle-treated mice.
An important aspect of the Ins1 Cre mouse model is that its glucose homeostasis and body weight control are indistinguishable from those of control littermates. It has been reported that one of the rat insulin promoter (RIP)-driven Cre mouse lines [11] has a defect in glucose homeostasis [27], casting some doubt on the results of reports showing an impact of beta cell gene deletion using this recombinase line. We have not tested body weight and glucose homeostasis in cohorts of Ins1 CreERT2 mice. However, because both the Ins1 Cre and Ins1 CreERT2 mice were generated by the same knock-in strategy we anticipate similar effects. In the different transgenic mouse lines, such as RIP-Cre, functional differences may result from insertional mutagenesis caused by the random insertion of the transgene, usually as large concatemers. This can potentially lead to unwanted effects such as the formation of loss-of-function alleles, abnormalities in the activity of nearby genes, and unexpected expression pattern and strength of the transgene. The knock-in approach used here results in a well-defined targeted modification that is therefore more precise and less prone to induction of artefacts. Another theoretical consideration is that the Ins2 promoter is stronger than Ins1 [28] and could thus result in higher concentrations of Cre that are potentially deleterious. Importantly, our knock-in approach does not affect glucose homeostasis despite suppressing one Ins1 allele, which is also in agreement with previous studies reporting perfect glycaemic control in mice with deletion of single alleles of the Ins1 or Ins2 locus [29].
In summary, we have generated two new Cre deleter mice that can be used for beta cell-specific constitutive (Ins1 Cre) or inducible (Ins1 CreERT2) deletion of genes. The constitutive deleter mice induce deletion in nearly all beta cells from the immediate postnatal period to adulthood and they display normal control of body weight and glycaemia. The inducible deleter mice will be useful to test the impact of gene deletion in adult mice or at a defined time during development, although deletion does not occur in all beta cells. These two mouse strains are therefore very valuable new tools for beta cell research.
Abbreviations
- Cre:
-
Cre recombinase
- CreERT2:
-
Cre-oestrogen receptor fusion protein
- eYFP:
-
Enhanced yellow fluorescent protein
- Flp:
-
Flippase
- GFP:
-
Green fluorescent protein
- Neo:
-
Neomycin
- RIP:
-
Rat insulin promoter
References
Thorens B (2013) The required beta cell research for improving treatment of type 2 diabetes. J Int Med 274:203–214
Kulkarni RN, Zisman A (2003) Lessons for human diabetes from experimental mouse models. Curr Diabetes Rep 3:168–175
Leiter EH (2002) Mice with targeted gene disruptions or gene insertions for diabetes research: problems, pitfalls, and potential solutions. Diabetologia 45:296–308
Patti M-E, Kahn CR (1996) Lessons from transgenic and knockout animals about noninsulin-dependent diabetes mellitus. Trends Endocrinol Metab 7:311–319
Moller DE (1994) Transgenic approaches to the pathogenesis of NIDDM. Diabetes 43:1394–1401
Aston-Mourney K, Subramanian SL, Zraika S et al (2013) One year of sitagliptin treatment protects against islet amyloid-associated beta-cell loss and does not induce pancreatitis or pancreatic neoplasia in mice. Am J Physiol Endocrinol Metab 305:E475–E484
Hu He KH, Lorenzo PI, Brun T et al (2011) In vivo conditional Pax4 overexpression in mature islet beta-cells prevents stress-induced hyperglycemia in mice. Diabetes 60:1705–1715
Martin M, Hauer V, Messmer M, Orvain C, Gradwohl G (2007) Transcription factors in pancreatic development. Animal models. Endocr Dev 12:24–32
Hennige AM, Burks DJ, Ozcan U et al (2003) Upregulation of insulin receptor substrate-2 in pancreatic beta cells prevents diabetes. J Clin Invest 112:1521–1532
Christen U, von Herrath MG (2002) Transgenic animal models for type 1 diabetes: linking a tetracycline-inducible promoter with a virus-inducible mouse model. Transgenic Res 11:587–595
Postic C, Shiota M, Niswender KD et al (1999) Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem 274:305–315
Hanahan D (1985) Heritable formation of pancreatic beta cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature 315:115–122
Magnuson MA, Osipovich AB (2013) Pancreas-specific Cre driver lines and considerations for their prudent use. Cell Metab 18:9–20
Indra AK, Warot X, Brocard J et al (1999) Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res 27:4324–4327
Herrera PL (2000) Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development 127:2317–2322
Wicksteed B, Brissova M, Yan W et al (2010) Conditional gene targeting in mouse pancreatic ss-Cells: analysis of ectopic Cre transgene expression in the brain. Diabetes 59:3090–3098
Hamilton-Williams EE, Palmer SE, Charlton B, Slattery RM (2003) Beta cell MHC class I is a late requirement for diabetes. Proc Natl Acad Sci U S A 100:6688–6693
Deltour L, Leduque P, Blume N et al (1993) Differential expression of the two nonallelic proinsulin genes in the developing mouse embryo. Proc Natl Acad Sci U S A 90:527–531
Devaskar SU, Singh BS, Carnaghi LR, Rajakumar PA, Giddings SJ (1993) Insulin II gene expression in rat central nervous system. Regul Pept 48:55–63
Srinivas S, Watanabe T, Lin CS et al (2001) Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1:4
Madisen L, Zwingman TA, Sunkin SM et al (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140
Tarussio D, Metref S, Seyer P et al (2014) Nervous glucose sensing regulates postnatal beta cell proliferation and glucose homeostasis. J Clin Invest 124:413–424
Solar M, Cardalda C, Houbracken I et al (2009) Pancreatic exocrine duct cells give rise to insulin-producing beta cells during embryogenesis but not after birth. Dev Cell 17:849–860
Mehran AE, Templeman NM, Brigidi GS et al (2012) Hyperinsulinemia drives diet-induced obesity independently of brain insulin production. Cell Metab 16:723–737
Nakamura K, Minami K, Tamura K, Iemoto K, Miki T, Seino S (2011) Pancreatic beta-cells are generated by neogenesis from non-beta-cells after birth. Biomed Res 32:167–174
Tamarina NA, Roe MW, Philipson L (2014) Characterization of mice expressing Ins1 gene promoter driven CreERT recombinase for conditional gene deletion in pancreatic beta-cells. Islets. doi:10.4161/isl.27685
Lee JY, Ristow M, Lin X, White MF, Magnuson MA, Hennighausen L (2006) RIP-Cre revisited, evidence for impairments of pancreatic beta-cell function. J Biol Chem 281:2649–2653
Hay CW, Docherty K (2006) Comparative analysis of insulin gene promoters: implications for diabetes research. Diabetes 55:3201–3213
Leroux L, Desbois P, Lamotte L et al (2001) Compensatory responses in mice carrying a null mutation for Ins1 or Ins2. Diabetes 50(Suppl 1):S150–S153
Acknowledgements
We thank V. Grau (IDIBAPS, Barcelona, Spain) for expert technical assistance. The Ins1 Cre and Ins1 CreERT2 mice were generated by Genoway and we thank K. Thiam (Genoway, Lyon, France) for discussing the targeting strategy.
Funding
This project was funded by a framework programme 6 project of the European Union (EuroDia; LSHM-CT-2006-518153) and by grants from the Swiss National Science Foundation (no. 3100A0B-128657) to BT and Ministerio de Economía y Competitividad (SAF2011-27086) to JF. JF is a Wellcome Trust Senior Investigator (WT101033).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
BT and JF conceived the project, supervised its realisation, analysed the data and wrote the manuscript. DT, MAM, MR and EH performed the experiments, analysed the data and contributed to writing of the manuscript. BT and JF are the guarantors of this work. All authors have approved the final version.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM Fig. 1
(PDF 329 kb)
ESM Fig 2
(PDF 430 kb)
ESM Table 1
(PDF 66 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Thorens, B., Tarussio, D., Maestro, M.A. et al. Ins1 Cre knock-in mice for beta cell-specific gene recombination. Diabetologia 58, 558–565 (2015). https://doi.org/10.1007/s00125-014-3468-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-014-3468-5