Abstract
Autophagy is a lysosomal degradation pathway recycling intracellular long-lived proteins and damaged organelles, thereby maintaining cellular homeostasis. In addition to inflammatory processes, autophagy has been implicated in the regulation of adipose tissue and beta cell functions. In obesity and type 2 diabetes autophagic activity is modulated in a tissue-dependent manner. In this review we discuss the regulation of autophagy in adipose tissue and beta cells, exemplifying tissue-specific dysregulation of autophagy and its implications for the pathophysiology of obesity and type 2 diabetes. We will highlight common themes and outstanding gaps in our understanding, which need to be addressed before autophagy could be envisioned as a therapeutic target for the treatment of obesity and diabetes.
Similar content being viewed by others
Introduction
Autophagy, an evolutionarily conserved process, functions in trafficking cytosolic components to the lytic compartment of the cell for degradation [1]. Autophagy is mediated by double-membrane vesicles, called autophagosomes, which sequester cytosolic constituents, including whole organelles, and fuse with lysosomes where the sequestered material is degraded and recycled. This is tightly regulated by a family of proteins encoded by autophagy genes (ATGs). More than 30 proteins have been identified in yeast [2], and for most, human orthologues exist. Autophagy proteins can be functionally grouped into those participating in: (1) membrane nucleation, elongation and formation of the double-membrane vesicle; (2) fusion with the primary lysosome to form autophagolysosomes; and (3) acid hydrolysis to degrade the vesicle content.
To survive scarcity of exogenous energy sources and/or amino acids, cells activate autophagy to ensure nutrient supply from endogenous sources. In addition, autophagy degrades misfolded proteins and damaged organelles, and is thus essential for cellular 'housekeeping' and survival under conditions of energy surplus. In line with this, autophagy is necessary in cells exposed to stress, including low-grade inflammation, which develops in obesity and diabetes [3, 4]. This is particularly relevant for metabolically active tissues such as beta cells and adipose tissue.
Beyond its ubiquitous role in maintaining cellular homeostasis, autophagy also regulates tissue-specific functions and modulates metabolism systemically. As an example, autophagy has been implicated in protein breakdown in the liver, required for gluconeogenesis and maintenance of glucose homeostasis while fasting [5]. A specific form of autophagy, termed lipophagy, regulates intracellular lipid stores via autophagosome-mediated triacylglycerol hydrolysis [6].
In addition to nutrients, hormones, most notably insulin and glucagon, are key regulators of autophagy [7, 8]; insulin inhibits, whereas glucagon stimulates, autophagy. Autophagy's tight regulation by both nutrient availability and hormones, along with its roles in whole-body metabolism and cellular adaptation to stress, hint at its involvement in the pathophysiology of common metabolic disorders, such as obesity and type 2 diabetes. While evidence is mounting for dysregulated autophagy in obesity and diabetes, it is still not settled whether ‘primary’ alterations in autophagic genes contribute to the development of obesity or type 2 diabetes, as has been established for certain other diseases, including Crohn's disease [9].
Impaired autophagy may greatly affect cellular homeostasis through the accumulation of damaged intracellular products leading to dysfunction or even cell death, potentially contributing to diabetes pathogenesis. Conversely, low-grade inflammation, oxidative and endoplasmic reticulum (ER) stress, insulin resistance, and adipose tissue hypoxia, all implicated in obesity and diabetes, may activate autophagy [10]. This could either serve to restore homeostasis and limit cellular dysfunction, or conversely, could contribute to pathogenesis, as aberrant activation of autophagy can result in excessive self-digestion and cell death [11]. Thus, dysregulated autophagy is a double-edged sword, with both activation and inhibition potentially contributing to pathogenesis. Another layer of complexity is the fact that autophagy is regulated in a highly tissue- and cell-type specific manner, even in the same organism [12]; this may serve to regulate cell-type specific functions.
Recent reviews have summarised the literature on autophagy in metabolic disorders [4, 7, 13–16]. The aim of the present article is not to provide an exhaustive review of the role of autophagy in these pathologies, but to highlight major concepts and important questions, controversies and challenges related to the regulation and physiological functions of autophagy in obesity and diabetes. We will focus on autophagy in adipose tissue and the beta cell, not only because of their central role in the pathogenesis of diabetes and obesity, but also because of the differential, tissue-specific dysregulation of autophagy observed within these tissues. We will highlight common themes and outstanding questions that need to be addressed before autophagy could be envisioned as a therapeutic target for the treatment of obesity and diabetes.
Assessment of autophagy in obesity and diabetes
The assessment of autophagy is frequently based on static quantifications of autophagosome and autolysosome numbers by immunofluorescence-based detection of the autophagosome membrane-associated LC3 (LC3-ΙΙ), which appears as punctae. Alternatively, ultrastructural morphology of autophagic vesicles is determined by electron microscopy, without dynamic functional assessment. Dynamic assessment is nevertheless crucial, since some autophagic components are degraded as part of the process. Hence, methodological issues involved in determining autophagic activity can lead to paradoxical or erroneous conclusions. For example, an increased number of autophagosomes may indicate either their increased formation (stimulated autophagy), or inhibition of autophagosome–lysosome fusion and/or maturation (inhibited autophagic flux). Thus, assessment of ‘autophagic flux’ is essential for the correct interpretation of autophagic activity. Several assays have been developed to assess autophagic flux [11], including measurement of global long-lived protein degradation using pulse–chase experiments and by measuring the accumulation of the autophagosome marker LC3-ΙΙ in the presence of lysosomal enzyme inhibitors. Greater accumulation of LC3-ΙΙ in response to inhibition of lysosomal degradation indicates that autophagic flux is enhanced. LC3-ΙΙ accumulation is analysed by western blot or by immunofluorescence and quantification of LC3 punctae. An additional common approach is the measurement of the abundance of proteins that undergo lysosomal degradation, such as p62/sequestosome 1 (SQSTM1). Decreased steady-state expression of p62/SQSTM1 suggests increased autophagic flux.
However, the specificity of these assays (to distinguish autophagy from other lysosomal-related processes) is low, and no accepted ‘gold standard’ currently exists. Therefore, adequate assessment of autophagy still typically requires the use of multiple approaches, as previously described [11, 13]. In vivo assessment of autophagic flux is difficult and is particularly challenging in humans, hampering the advancement of our knowledge on the autophagic process. Hence, the development of sensitive and accurate assays to monitor autophagy in a tissue-specific manner is greatly needed.
The role of autophagy in adipose dysfunction and inflammation
Autophagy is involved in several aspects of adipocyte biology and may modulate immune cell function [17]. The following sections will discuss the role of autophagy in adipogenesis and immune cell biology, its metabolic regulation and its putative impact on adipose tissue dysfunction and inflammation in obesity and type 2 diabetes (Figs 1 and 2).
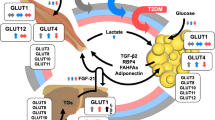
Regulation of autophagy in lean and obese adipose tissue and its impact on adipocytes. In the lean state (a), autophagy is regulated by the nutrient sensors mTORC1 and AMPK, which inhibit and activate the process, respectively. Similarly to nutrients, insulin also stimulates mTORC1, leading to inhibition of autophagy. Autophagy promotes white adipose tissue differentiation, whereas inhibition of autophagy induces a ‘browning’ phenotype of the adipose tissue. In obesity (b), stimulation of autophagy may have differential effects on adipocyte function and survival. Nutrient (glucose and NEFA) overload stimulates mTORC1 and inhibits AMPK, thereby attenuating autophagy. By contrast, ER stress, hypoxia and inflammation, conditions that are commonly observed in obese adipose tissue, induce insulin resistance, leading to inhibition of mTORC1 and consequently to stimulation of autophagy. Autophagy may improve adipocyte function by eliminating misfolded proteins and damaged organelles and attenuating the proinflammatory response of obesity. On the other hand, excessive stimulation of autophagy may enhance adipocyte energy storage and promote ‘self-digestion’ and consequently cell death
Regulation of autophagy in lean and obese adipose tissue and its impact on adipose tissue macrophages. In the lean state (a), macrophages exhibit an anti-inflammatory phenotype. Autophagy is inhibited by mTORC1 and activated by AMPK. Stimulated autophagy curtails proinflammatory responses by reducing proinflammatory gene expression and inflammasome activity, thereby inhibiting the processing and activation of IL-1β along with enhanced degradation, hence limiting IL-1β availability for inflammasome-dependent processing. In obesity (b), the inflammatory trait of the macrophage is augmented. LPS and inflammatory cytokines activate autophagy, whereas nutrient (glucose and NEFA) overload stimulates mTORC1 and inhibits AMPK, resulting in attenuation of autophagy. Autophagy restrains the proinflammatory response by limiting proinflammatory gene expression and inflammasome activation. In addition, lipophagy may attenuate the inflammatory response by promoting the availability of NEFA for oxidative metabolism, a key metabolic pathway that drives an anti-inflammatory trait. The biogenesis of lysosomes is increased in macrophages that populate obese adipose tissue, which may promote lipid trafficking within the macrophages
Essential role for autophagy in adipocyte development and function
Autophagy is required for lipid storage and components of the autophagic machinery, for example ATG5 and ATG7 are required for white adipocyte cell differentiation [18]. Mouse embryonic fibroblasts (MEFs) derived from Atg5 −/− (autophagy-deficient) animals exhibit reduced adipogenesis [18], with an initiation of the process and triacylglycerol accumulation, but failure to complete adipocyte maturation. Consistently, adipogenesis was impaired in vivo when ATGs were genetically targeted [19, 20]; white adipose tissue depots were smaller in Atg7 −/− animals, suggestive of a defect in adipocyte differentiation. Moreover, adipocyte-specific knockdown of ATG7 resulted in the development of adipose tissue resembling brown fat, with multi-loculated adipocytes and abundant mitochondria, indicating increased oxidative capacity [20]. The precise mechanism(s) by which autophagy regulates adipogenesis is unknown. An intriguing hypothesis is that autophagosomes with membranes originating from potentially different intracellular organelles serve to mobilise membranes within the cell, thereby facilitating the reorganisation of cytoplasmic components, which is thought to be a requirement for adipogenesis [21]. In addition, autophagy has been reported to increase the stability of peroxisome proliferator-activated receptor (PPAR)γ2, the master regulator of adipocyte differentiation and adipogenesis [22].
Intriguingly, impaired adipogenesis was also observed in animals with skeletal muscle-specific deletion of Atg7 [23]. This suggests that alterations in autophagic activity can modulate inter-organ crosstalk, a notion also supported by the observation of impaired lipolysis when autophagy was genetically disrupted in hypothalamic pro-opiomelanocortin (POMC) neurons [24].
Altogether, functional autophagy appears to support adipocyte development and differentiation. Hence, any primary/developmental disturbance in autophagy may affect adipose tissue mass and homeostasis.
Autophagy and inflammation
The interaction between autophagy and the innate immune system is well established [17]; however, the molecular mechanisms involved are still elusive. Since clearance of many pathogens by immune cells critically depends on autophagy, it is not surprising that toll-like receptor (TLR) activation stimulates autophagy [25]. Obesity has been proposed as a low-grade ‘metabolic endotoxaemia’ state [26]. Lipopolysaccharide (LPS), the most common endotoxin, is included in the cell wall of Gram-negative bacteria in the gut. In obesity, intestinal permeability is increased, leading to elevated circulating endotoxin levels [26]. Inasmuch as both adipocytes and adipose tissue inflammatory cells express TLR4 [27], its stimulation by circulating LPS may activate autophagy. TLR-stimulated autophagy may function as a negative feedback mechanism aimed at restricting inflammation. In line with this hypothesis, deficiency of the autophagic protein ATG16L1 augmented IL-1β processing upon TLR4 stimulation [28]. Further, autophagy reduced the expression and subsequent secretion of specific proinflammatory cytokines, including IL-1β. In addition, pro-IL-1β is degraded in autophagosomes, thus limiting its availability for inflammasome-dependent activation [29]. Finally, several studies indicate that autophagy directly inhibits inflammasome activation [30], which in turn was suggested to regulate adipose tissue inflammation [31]. Collectively, these results imply that autophagy may restrain the innate immune response, thus curtailing adipose tissue inflammation and dysfunction. However, there is also evidence challenging this notion, as stimulated IL-1β secretion may engage activated autophagy and/or elements of the autophagic machinery [32].
Metabolic regulation of adipose tissue inflammation and autophagy
Recent advances in immunology have revealed that the intracellular metabolism of innate immune cells determines their activation [33]. More specifically, enhanced glucose utilisation through glycolysis is crucial for an adequate proinflammatory immune response. Although, from an energetic standpoint, glycolysis-mediated ATP production is inefficient compared with oxidative phosphorylation, it promotes the synthesis of various macromolecules, which in turn may increase cytokine production [33]. Whereas activation of glycolysis favours the proinflammatory response of an immune cell, enhancement of oxidative phosphorylation drives anti-inflammatory actions. Indeed, M2 alternatively activated (anti-inflammatory) macrophages exhibit increased reliance on oxidative phosphorylation over glycolysis for energy production [34].
Glycolysis and oxidative phosphorylation are regulated by mammalian target of rapamycin complex 1 (mTORC1) and AMP-activated protein kinase (AMPK), two major nutrient-sensing kinases. Under conditions of energy shortage, mTORC1 is inhibited whereas AMPK is activated, thereby inhibiting anabolic pathways and stimulating ATP-producing processes, including mitochondrial fatty-acid oxidation. Shifting from glycolytic metabolism to oxidative phosphorylation may attenuate the proinflammatory response [33]. Indeed, both inhibition of mTORC1 by rapamycin [18] and activation of AMPK by 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) [35] exert anti-inflammatory responses.
Importantly, mTORC1 and AMPK are also key regulators of autophagy. mTORC1 prevents initiation of autophagy by phosphorylating ATG13, thereby blocking its binding to ATG1 (also called ULK1 in mammals) and the formation of the ATG1–ATG13–ATG17 complex [36]. Thus, inhibition of mTORC1 during starvation, or pharmacologically by agents such as rapamycin, stimulates the initiation of autophagy. AMPK stimulates autophagy by inhibiting mTORC1 and by phosphorylating ULK1 [37]. In macrophages (and probably also in other immune cells) activation by AMPK-induced lipid droplet autophagy (lipophagy) provides NEFA as substrates for oxidative phosphorylation [1], curtailing the proinflammatory state. In contrast, mTORC1-mediated inhibition of autophagy, which eventually may promote glycolysis, elicits a proinflammatory response. Hence, variations in autophagic activity may greatly impact on the metabolic status of immune cells, thereby altering the inflammatory trait of the adipose tissue.
Aberrant regulation of adipose tissue autophagy in obesity and diabetes
In adult mammals, the adipose tissue is comprised of multiple cell types, including adipocytes, stromal-vascular cells, fibroblasts, adipocyte precursors and various inflammatory cells. In obesity, adipocytes may constitute a significantly smaller fraction of the cell types comprising adipose tissue, and therefore studying biological processes in isolated adipocytes is important. Ost et al were the first to show that autophagosome content is increased in isolated adipocytes derived from obese and diabetic humans [38]. Furthermore, an increase in the autophagic flux was demonstrated. These findings were confirmed in whole adipose tissue derived from the omentum or subcutaneous depot of obese patients undergoing bariatric surgery [39–41]. Assessment of autophagic flux by confocal microscopy using whole-tissue fragments suggested that elevated autophagy was present in both adipocytes and non-adipocytes [39].
In contrast to the findings in human adipocytes and adipose tissue, in rodents autophagy was reduced both in vitro and in adipose tissue of animals fed a high-fat diet (HFD) for 16 weeks [42]. These conflicting findings may be explained by species differences or by specific experimental conditions (e.g. different fasting periods prior to autophagy assessment). Yet, overall, in vivo determinations of adipose autophagic flux are required to demonstrate unequivocally whether an elevated number of autophagosomes in adipose tissue in obesity actually corresponds to activated or inhibited autophagy. Despite these technical obstacles, the current accepted view is that autophagy is enhanced in obese adipose tissue, although the underlying mechanisms are not completely understood.
Insulin is an anabolic hormone that functions as a potent inhibitor of autophagy. Experimental data and mathematical modelling showed that, in type 2 diabetes, expansion of the adipose tissue leads to insulin resistance impairing insulin activation of mTORC1 [38, 43]; this may readily explain autophagy activation observed in adipose tissue of obese individuals, consistent with the observation of Ost et al [38]. In addition, inflammation, ER stress and hypoxia, conditions that are commonly observed in adipose tissue during obesity, also inhibit mTORC1 [44], thus further promoting autophagy. However, mTORC1 can be activated by glucose, amino acids and fatty acids, which are overabundant in diabetes [44]. The notion that autophagy is stimulated in obese adipose tissue may suggest that the stimulatory effects of insulin resistance, stress, inflammation and/or hypoxia prevail over the expected inhibitory effect of mTORC1 activation by nutrient overload. Interestingly, following bariatric surgery in obese diabetic patients, adipose tissue autophagy decreases despite continued ER stress. This suggests that other regulators of adipose tissue autophagy, such as insulin resistance, may dominate the control of adipose autophagy in response to weight loss [41].
Given the high cell-type specificity in the regulation of autophagy, future studies should aim at deciphering the level of autophagic activation in the various cell types that comprise the adipose tissue, including immune, endothelial and adipocyte precursor cells.
How might altered adipose tissue autophagy affect adipose function?
Based on currently available knowledge, it is not clear whether stimulation of autophagy in adipose tissue during obesity is deleterious or beneficial. Associatively, activated autophagy is apparent in whole adipose tissue in obesity when insulin resistance develops, but before cardio-metabolic morbidity ensues [39]; however, this does not prove causality. In mice, disruption of autophagy in adipocytes decreased body weight and enhanced insulin sensitivity. This was accompanied by a decrease in adipose tissue inflammation [19]. Thus, whether this protective effect was a direct consequence of the autophagic inhibition, or an indirect consequence of reduced body weight gain is difficult to establish. Moreover, in these models, adipocyte-specific interference with autophagy was achieved genetically using an adipocyte promoter, thereby affecting adipocytes from their early development onwards. Whether modulation of autophagy in fully mature adipocytes, secondary to obesity, would have similar effects remains to be established. Inhibited autophagy leads to adipose tissue browning and energy dissipation; conversely, activated autophagy might serve to functionally adapt adipocytes to energy storage in response to chronic over-nutrition (‘whitening’). Indeed, if activated autophagy leads to a decrease in mitochondrial mass and/or its oxidative capacity, it would enable adipocytes to store extra calories as triacylglycerol more efficiently. Finally, beyond direct regulation of energy storage and cell survival mechanisms, excessive autophagy may also constitute a cell death mechanism. Indeed, adipocyte cell death has been implicated as a major pathogenic mechanism in the development of adipose tissue inflammation during obesity [45]. Hence, activated autophagy could potentially contribute to adipocyte cell death. Collectively, the potential role of autophagy in regulating the inflammatory tone (as mentioned above) awaits further experimental elucidation.
Frequently, autophagy is viewed as an adaptive response, so its activation in obesity may protect cells from further metabolic stress and inflammation. As such, inhibiting autophagy may be deleterious, depriving the tissue of an essential protective mechanism. Obese adipose tissue is exposed to hypoxia and ER stress [46], which might lead to accumulation of misfolded proteins [47] that can be partly eliminated by autophagy. Consistent with this paradigm, inhibition of autophagy led to increased ER stress and subsequently promoted adipose tissue inflammation [40, 42].
The complex regulation of autophagy in adipose tissue and the possible paradoxical effects of modulating autophagy in obesity emphasises the challenge of elucidating its role in the pathophysiology of obesity and type 2 diabetes, and how manipulating the process might be utilised for therapeutic purposes.
Effects of dysregulated autophagy in the hypothalamus on obesity
The impact of dysregulated autophagy in obesity and diabetes is further complicated by inter-organ communications that may affect whole-body metabolism. One particularly intriguing example is the hypothalamus, which functions as a hub that integrates metabolic and hormonal cues to regulate food intake and energy expenditure, thereby modulating lipid metabolism and glucose homeostasis [48, 49]. Obesity might induce hypothalamic dysfunction by stimulating inflammation through the inhibitor of κβ (IKKβ)/nuclear factor-κB (NF-κβ) pathway leading to hypothalamic resistance to insulin and the satiety hormone, leptin [50]. Intriguingly, intra-cerebroventricular injection of TNFα has been shown to impair insulin secretion [51]. Hence, hypothalamic dysfunction may play a role in the pathophysiology of obesity and diabetes.
Interestingly, hypothalamic inhibition of autophagy using ATG7 siRNA-mediated knockdown resulted in increased energy consumption and reduced energy expenditure, leading to impaired adipose lipolysis, exacerbation of obesity and whole-body insulin resistance in response to HFD feeding [24, 52].
Moreover, impaired hypothalamic autophagy has been observed during HFD-induced obesity [52]. Similarly to the situation with adipose tissue, impaired hypothalamic autophagy may elicit a local inflammatory response probably leading to hypothalamic dysfunction. Altogether, defective autophagy might cause hypothalamic inflammation and dysfunction, leading to obesity, systemic insulin resistance and probably beta cell dysfunction. This intriguing hypothesis awaits further experimental confirmation.
Autophagy in beta cell physiology and diabetes
In obesity, the beta cell adapts to insulin resistance by increasing insulin production and secretion [53]. The life-long stimulus for the beta cell to secrete large amounts of insulin for maintaining euglycaemia is associated with an increased protein-folding burden in the ER. This may lead to accumulation of misfolded proteins, most importantly proinsulin, resulting in ER stress [54, 55]. This, together with oxidative stress elicited by excessive mitochondrial generation of reactive oxygen species, leads to beta cell dysfunction [56, 57]. The latter is the driving force for progression from obesity to diabetes. Moreover, beta cell dysfunction is the main cause for deterioration of glycaemic control in diabetes over time. Type 2 diabetes is accompanied by elevated NEFA, hyperglycaemia and inflammation. Each of these factors increases cellular stress, thus generating a feed-forward vicious cycle that impinges on beta cell function, and may induce apoptosis and probably beta cell dedifferentiation [58].
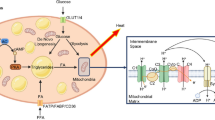
Autophagy may protect the stressed beta cell by eliminating damaged organelles (mitochondria [mitophagy] and ER [reticulophagy] [59, 60]) and/or misfolded proteins, notably proinsulin. Accumulating data support this notion and suggest that lysosomal degradation pathways, including autophagy and crinophagy, are important for beta cell homeostasis both in physiology and in diabetes (Fig. 3).
The role(s) of autophagy in beta cell physiology and dysfunction in diabetes. Beta cells are prone to the inter-related oxidative and ER stresses, even in the normoglycaemic state (a). Autophagy may prevent cellular stress by eliminating misfolded proteins, including proinsulin, and probably dysfunctional organelles. For example, through mitophagy damaged mitochondria are efficiently removed and thereby oxidative stress is restrained. In addition, the autophagic machinery and other lysososomal degradation pathways decrease the insulin granule pool and may restrain insulin secretion by lysosomal lipid degradation (lipophagy). The metabolic milieu of obesity and diabetes may have differential effects on autophagic activity in the beta cell (b). Hyperglycaemia, elevated NEFA and probably hIAPP may inhibit autophagy either by stimulating mTORC1 (glucose and NEFA) or by inhibiting lysosomal acidification (NEFA and hIAPP). In contrast, ER stress may stimulate autophagy, which in turn improves the beta cell adaptation to stress. The regulation of beta cell autophagy in diabetes and its impact on beta cell function and survival is controversial (see text for details)
Physiological roles of crinophagy and autophagy
Pioneering studies by Orci et al showed that insulin granules are regulated by lysosomal degradation through crinophagy and autophagy [61]. In crinophagy, the secretory granule membrane fuses with the membrane of a large vacuolar, lysosomal compartment to generate a crinophagic body, within which the insulin granule content is degraded. Insulin granules may also reach lysosomes via autophagosomes that engulf cytosolic components containing secretory granules (macroautophagy), or via lysosomal engulfment and swallowing of a single granule (microautophagy). Insulin is relatively resistant to degradation in the acidic environment of the lysosome; its degradation is much slower than that of C-peptide or proinsulin [61, 62]. This underlies the assumption that insulin degradation plays a minor role in the regulation of insulin homeostasis and beta cell function [62]. On the other hand, crinophagic activity and insulin degradation are modulated by glucose [63], suggesting that this process is dynamic and tightly regulated: at low glucose, insulin degradation in the beta cell increases, whereas stimulation of insulin secretion at high glucose is associated with inhibition of its degradation [63, 64]. The biological rationale for insulin granule degradation at low glucose remains unclear: increased insulin content through accumulation of unsecreted granules is not expected to cause hypoglycaemia, since exocytosis is inhibited under these conditions; in addition, in response to glucose stimulation only a minute fraction of the total insulin granule pool is released. It is therefore possible that elimination of ‘old’ granules, while energetically costly, serves to increase the intracellular amino acid pool.
In addition to the putative role of autophagy in insulin granule degradation, stimulation of autophagy in the post-absorptive state may maintain cellular homeostasis by eliminating dysfunctional organelles. As an example, in beta cells mitochondria undergo rapid cycles of fusion and fission; the latter is associated with generation of depolarised mitochondria that are then eliminated by autophagy [65]. Stimulating autophagy during fasting may prevent oxidative injury by enhancing the clearance of damaged and/or depolarised mitochondria, which have accumulated during ‘hyperactive’ (postprandial) periods. This is largely reminiscent of the proposed role of augmented mitophagy during fasting or in response to glucagon in the liver [66].
Intriguingly, inhibition of insulin secretion either by pharmacological means (diazoxide) [67] or by interfering with the insulin granule secretory machinery (Rab3a knockout mice [68] or mammalian uncoordinated [Munc]-18-1 depletion [69]) all stimulate insulin degradation via autophagy-related processes, thereby maintaining a stable intracellular insulin content. These findings suggest a close interaction between insulin secretion and autophagy/crinophagy.
Newly synthesised insulin granules are preferentially secreted in response to glucose stimulation [70], whereas old insulin granules are more likely to be degraded by crinophagy; hence, it was postulated that insulin granule degradation is a relatively long-term homeostatic mechanism for maintaining constant insulin stores [71]. Why maintaining a constant store of newly formed insulin granules should be important for optimal beta cell function is not clear, since the insulin content is always several orders of magnitude greater than the amount secreted. It is likely that autophagy functions as a quality control process by eliminating aged secretory granules, and is probably required for the maintenance of beta cell homeostasis. Recently, it was suggested that lysosomal lipid degradation (lipophagy) negatively regulates glucose-stimulated insulin secretion by depletion of substrate for non-lysosomal neutral lipases that regulate insulin secretion [72]. Thus, autophagy–lysosomal degradation may affect both insulin content and secretion.
There are still important questions that need to be addressed regarding the role of autophagy in insulin secretion and degradation: how does the beta cell sense insulin content or insulin granule number and age? How do glucose and insulin regulate insulin granule degradation? What governs and coordinates insulin degradation? Recent reports showed that SNAP receptor (SNARE) proteins play an important role in the regulation of autophagy [73]. SNARE proteins are involved both in insulin granule exocytosis and autophagosome and/or lysosome trafficking. It is possible that inhibition of insulin secretion, for example in fasting, may increase the availability of such proteins, thereby promoting autophagy.
Effects of inhibited autophagy on beta cell function and adaptation to obesity
Interfering with autophagy functionally disrupts beta cell function: beta cell specific Atg7 knockout mice exhibited glucose intolerance without developing full-blown diabetes [74, 75]. This resulted from insulin deficiency and impairment of beta cell function, evident by decreased glucose-stimulated Ca2+ influx and ATP production, paralleled by reduced basal and stimulated insulin secretion. Polyubiquitinated protein aggregates accumulate in the cytosol of autophagy-deficient beta cells [74, 75]. Electron microscopy shows vacuolar degeneration of the beta cells, along with swelling of mitochondria and cisternal expansion of the ER, indicating that impaired autophagy induces cellular stress. Beta cell apoptosis was increased and proliferation reduced, resulting in decreased beta cell mass.
Surprisingly, the expression of genes involved in cellular protection against ER stress, including anti-oxidants and unfolded protein response (UPR) genes, were all reduced in autophagy-deficient beta cells [76]. This rendered autophagy-impaired beta cells hyper-susceptible to apoptosis in response to ER stress. Furthermore, autophagy deficiency prevented the compensatory increase in beta cell mass in response to HFD and in ob/ob mice, resulting in further deterioration of glucose tolerance [75, 76]. These studies convincingly show that basal autophagy is not only indispensable for maintaining beta cell mass and function, but is also required for beta cell compensation to obesity-induced insulin resistance. Thus, impairment of autophagy in beta cells may contribute to progression from obesity to diabetes.
Is beta cell autophagy impaired in diabetes?
Reminiscent of the situation in adipose tissue, an increased number of autophagosomes and autophagolysosomes was observed in the beta cells of different models of type 2 diabetes, including ob/ob and db/db mice [75, 76] and Akita mice [10]. In Zucker diabetic fatty rats and in insulinoma cells, hyperglycaemia and oxidative stress led to the accumulation of polyubiquitinated protein aggregates that were degraded by autophagy [77]. Importantly, beta cells of human type 2 diabetic patients showed a massive overload of autophagic vacuoles that associated with beta cell death, without nuclear condensation, which was referred to as autophagy-associated cell death [78]. However, these morphological changes do not imply that beta cell death resulted from ‘hyperactive’ autophagy. Autophagic cell death should be defined on the basis of strict criteria, including demonstrating that autophagy is increased and that its inhibition decreases cell death. This was not systematically assessed, thus preventing any definitive conclusion on whether autophagy was inhibited or activated in different diabetes models and in human type 2 diabetes, and, consequently, whether there is any impact on beta cell function and survival. In Akita mice, a model of proinsulin misfolding-induced diabetes, autophagic flux was moderately increased in islets and in an Akita beta cell line [10]; this was demonstrated using multiple assays, but it is unclear whether these findings can be extrapolated to other models of diabetes. It is worth noting that autophagy-mediated cell death is a rare phenomenon, and its mere existence has been questioned [79]. Further studies are required to clarify the meaning of altered autophagy in diabetic beta cells.
Interestingly, HHEX/IDE was identified as a type 2 diabetes risk locus linked to impaired beta cell function. The Ide gene encodes a multifunctional protein implicated in proteasome activity and protein degradation [80]. Beta cells of Ide knockout mice exhibited decreased glucose-stimulated insulin secretion, together with reduced microtubule content and autophagic flux [80]. This may further support the presence of a link between autophagy and beta cell dysfunction in human type 2 diabetes.
How the diabetic environment (e.g. hyperglycaemia, elevated NEFA and human islet amyloid polypeptide [hIAPP], and inflammation) affects autophagy and its impact on beta cell function also remains controversial. Several studies have shown that NEFA inhibit the expression of lysosome enzymes [78], lysosomal acidification and autophagic flux [81]. Similarly, the amyloidogenic peptide hIAPP has been found to inhibit autophagy [82]. This might lead to accumulation of toxic hIAPP oligomers that exacerbate beta cell dysfunction and apoptosis. On the other hand, others have shown that both hyperglycaemia and elevated NEFA stimulate, rather than inhibit, autophagy [83–85]. Moreover, a recent report suggested that, in beta cells, stimulation of autophagy by NEFA induces a proinflammatory response through NLRP3 [85]. In addition, cytokines such as IL-1β may be secreted via an unconventional pathway involving autophagy, which may promote inflammation [32]. Autophagic stimulation of inflammation in beta cells (if reproducible) is in marked contrast with the common notion in adipose tissue, in which autophagy is proposed to attenuate the inflammatory response (see the section Autophagy and inflammation above). Such opposing effects of autophagy on inflammation may be explained by tissue specificity of the autophagy–inflammation crosstalk.
The apparent paradoxical findings in relation to autophagic activity in diabetes could result from methodological differences in monitoring autophagy, as discussed above. As an example, Danon disease (OMIM: 300257) results from lysosome-associated membrane protein-2 (LAMP-2) deficiency, which disrupts the maturation of autophagosomes and their fusion with lysosomes, hence impairing autophagy [86]. This results in autophagic vacuole accumulation in various tissues, including liver, pancreas, muscle and heart [87], resembling the findings in human diabetic islets [78]. Without monitoring the autophagic flux, these findings could be erroneously interpreted as stimulation of autophagy. Notably, LAMP-2 expression was decreased in human diabetic islets and in response to treatment with NEFA, along with decreased expression of the lysosome enzymes cathepsin B and cathepsin D [78], suggesting that autophagic flux might be impaired. Yet, beyond the technicalities and interpretation of experimental results, it appears that opposing forces governing autophagy operate in beta cells in diabetes, similarly to adipose tissue, potentially leading to inconsistent modulation of autophagy depending on the metabolic and biological environment of the cell. As an example, hyperglycaemia stimulates mTORC1 in beta cells [88, 89], which is expected to inhibit autophagy. By contrast, ER stress due to inflammation and accumulation of misfolded proinsulin may stimulate autophagy [90]. Thus, the degree of beta cell autophagy may vary depending on the intricate interactions between nutrient and stress signalling, which differentially influence the activity of mTORC1 and probably other regulators of autophagy.
Is stimulation of autophagy in the diabetic beta cells beneficial or deleterious?
The shortage of genetic and pharmacological tools to stimulate autophagy specifically in beta cells hampers the efforts to address this important question. The impact on beta cell function and survival of using rapamycin to stimulate autophagy has been studied in several models of beta cell stress and diabetes.
In the Akita model of diabetes, mice carry a mutation in one proinsulin allele, leading to the translation of an irreparably misfolded hormone. This mutant proinsulin is trapped in the ER, where it generates ER stress and marked reduction of insulin secretion, resulting in diabetes [91, 92]. Similar mutations were found in a rare form of human congenital diabetes, MIDY (mutant INS gene-induced diabetes of youth syndrome) [93]. Intriguingly, in Akita islets autophagy was not reduced; still, stimulation of autophagy by the mTORC1 inhibitors rapamycin or Torin1 (an mTOR kinase inhibitor) alleviated stress and prevented beta cell apoptosis, while inhibition of autophagy severely augmented cellular stress. The physiological relevance of these findings was demonstrated when treatment of diabetic Akita mice with rapamycin improved diabetes and increased pancreatic insulin content and secretion [10]. Similarly, rapamycin alleviated stress in an autophagy-dependent manner in insulin-secretion deficient beta cells derived from fetal mice [94] and in beta cells exposed to lipotoxicity [81]. It is noteworthy that this apparent protection induced by rapamycin is contrary to the common view of this drug as being diabetogenic [88, 95, 96]. These findings may suggest that stimulating autophagy by rapamycin is protective to beta cells only under certain stressful conditions, such as proinsulin misfolding.
Another diabetes model is the pancreatic and duodenal homeobox (Pdx)-1 deficient mouse, which exhibits reduced beta cell mass and insulin secretion. In these mice, beta cell autophagy is increased [97]. Interestingly, and in contrast to Akita mice, inhibition of autophagy by crossing the mice with Beclin1 haplo-insufficient mice improved beta cell function and increased beta cell mass after 1 week on HFD. However, this protective effect by autophagy inhibition was transient, and was no longer apparent after 7 weeks. Whether short-term inhibition of autophagy may improve beta cell function in other models of diabetes or is unique to this model remains to be elucidated.
Future development of pharmacological and genetic means to allow specific stimulation of autophagy is essential for testing the impact of modulating autophagy on diabetes development and progression and for clarifying the conditions under which stimulation or inhibition of autophagy would improve beta cell function and, therefore, might be beneficial.
Modulation of autophagy in adipose tissue and the beta cell: prospects for a new therapeutic target for diabetes?
Could modulation of autophagy in adipose tissue and beta cells become a therapeutic approach in type 2 diabetes? The complex regulation of autophagy in obesity and diabetes emphasises the need for additional knowledge before the targeting of autophagy could be considered as a therapeutic strategy. Given its apparent roles in normal physiology, it would appear that modulating autophagy could be used to treat patients with altered autophagic activity as part of their disease process, seeking to normalise, rather than to ‘artificially’ manipulate autophagy. Such an approach would seem to be both effective and safe. Unfortunately, biomarkers for in vivo monitoring of autophagic activity are currently not available, thus preventing identification of diabetic patients suffering from altered autophagic activity. In addition, targeted therapy to specific cells or tissues would be of great value, as modulation of autophagy may have differential effects depending on the specific cell type affected.
As discussed above, considerable evidence suggests that autophagy alleviates stress and attenuates inflammation in adipose tissue and probably also in beta cells; thus, stimulating autophagy seems an attractive approach to improve tissue adaptation to the inflammatory stress of obesity and diabetes. Common glucose-lowering medications, including metformin and thiazolidinediones, stimulate autophagy and prevent NEFA toxicity to beta cells [78, 98]. Consistent with stimulated autophagy, thiazolidinediones promote adiposity, attenuate adipose tissue inflammation and enhance insulin action in type 2 diabetes [99]; however, it is unknown whether these effects are mediated via these drugs' ability to stimulate autophagy.
On the other hand, several clinical trials suggest beneficial metabolic effects for anti-malaria agents that prevent acidification of lysosome-related compartments [100]. This would suggest that inhibition of autophagy or other lysosomal functions may turn out to be beneficial, probably due to inhibition of hepatic insulin degradation [101] or, as more recently proposed, by inhibiting lysosome-dependent lipolysis in adipose tissue macrophages [11].
In summary, additional studies are required to clarify the regulation of autophagy in diabetes and its impact on glucose homeostasis, stress and inflammation in different tissues. There is an urgent need for new tools to assess autophagic activity in vivo and for compounds that would specifically modulate autophagy. The recent development of a Beclin1-based autophagy-inducing peptide [102] may be important to advance our knowledge on the autophagic process in vivo. These studies will allow the identification of diabetic patients who may benefit from treatment with autophagy-modulating compounds, and will provide crucial information on when and how autophagy should be modulated as part of the treatment of patients with diabetes and/or obesity.
Abbreviations
- AMPK:
-
AMP-activated protein kinase
- ATG:
-
Autophagy gene
- ER:
-
Endoplasmic reticulum
- HFD:
-
High-fat diet
- hIAPP:
-
Human islet amyloid polypeptide
- LAMP-2:
-
Lysosome-associated membrane protein-2
- LPS:
-
Lipopolysaccharide
- mTORC1:
-
Mammalian target of rapamycin complex 1
- TLR:
-
Toll-like receptor
References
Ouimet M (2013) Autophagy in obesity and atherosclerosis: interrelationships between cholesterol homeostasis, lipoprotein metabolism and autophagy in macrophages and other systems. Biochim Biophys Acta 1831:1124–1133
Klionsky DJ (2007) Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 8:931–937
Kim KH, Lee MS (2013) Autophagy as a crosstalk mediator of metabolic organs in regulation of energy metabolism. Rev Endocr Metab Disord 15:11–20
Gonzalez CD, Lee MS, Marchetti P et al (2011) The emerging role of autophagy in the pathophysiology of diabetes mellitus. Autophagy 7:2–11
Ezaki J, Matsumoto N, Takeda-Ezaki M et al (2011) Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy 7:727–736
Singh R, Kaushik S, Wang Y et al (2009) Autophagy regulates lipid metabolism. Nature 458:1131–1135
Chen ZF, Li YB, Han JY et al (2011) The double-edged effect of autophagy in pancreatic beta cells and diabetes. Autophagy 7:12–16
Mortimore GE, Poso AR, Lardeux BR (1989) Mechanism and regulation of protein degradation in liver. Diabetes Metab Rev 5:49–70
Cadwell K, Liu JY, Brown SL et al (2008) A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456:259–263
Bachar-Wikstrom E, Wikstrom JD, Ariav Y et al (2013) Stimulation of autophagy improves endoplasmic reticulum stress-induced diabetes. Diabetes 62:1227–1237
Klionsky DJ, Abdalla FC, Abeliovich H et al (2012) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8:445–544
Maixner N, Kovsan J, Harman-Boehm I, Bluher M, Bashan N, Rudich A (2012) Autophagy in adipose tissue. Obes Facts 5:710–721
Yamada E, Singh R (2012) Mapping autophagy on to your metabolic radar. Diabetes 61:272–280
Fujitani Y, Ueno T, Watada H (2010) Autophagy in health and disease. 4. The role of pancreatic beta-cell autophagy in health and diabetes. Am J Physiol Cell Physiol 299:C1–C6
Choi AM, Ryter SW, Levine B (2013) Autophagy in human health and disease. N Engl J Med 368:651–662
Rubinsztein DC, Codogno P, Levine B (2012) Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov 11:709–730
Levine B, Mizushima N, Virgin HW (2011) Autophagy in immunity and inflammation. Nature 469:323–335
Baerga R, Zhang Y, Chen PH, Goldman S, Jin S (2009) Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy 5:1118–1130
Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S (2009) Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A 106:19860–19865
Singh R, Xiang Y, Wang Y et al (2009) Autophagy regulates adipose mass and differentiation in mice. J Clin Invest 119:3329–3339
Zhang Y, Zeng X, Jin S (2012) Autophagy in adipose tissue biology. Pharmacol Res 66:505–512
Zhang C, He Y, Okutsu M et al (2013) Autophagy is involved in adipogenic differentiation by repressesing proteasome-dependent PPARγ2 degradation. Am J Physiol Endocrinol Metab 305:E530–E539
Kim KH, Jeong YT, Oh H et al (2013) Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med 19:83–92
Kaushik S, Arias E, Kwon H et al (2012) Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Rep 13:258–265
Martinez J, Verbist K, Wang R, Green DR (2013) The relationship between metabolism and the autophagy machinery during the innate immune response. Cell Metab 17:895–900
Piya MK, Harte AL, McTernan PG (2013) Metabolic endotoxaemia: is it more than just a gut feeling? Curr Opin Lipidol 24:78–85
Schaffler A, Scholmerich J (2010) Innate immunity and adipose tissue biology. Trends Immunol 31:228–235
Saitoh T, Fujita N, Jang MH et al (2008) Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456:264–268
Harris J, Hartman M, Roche C et al (2011) Autophagy controls IL-1β secretion by targeting pro-IL-1β for degradation. J Biol Chem 286:9587–9597
Plantinga TS, Joosten LA, van der Meer JW, Netea MG (2012) Modulation of inflammation by autophagy: consequences for Crohn's disease. Curr Opin Pharmacol 12:497–502
Stienstra R, Tack CJ, Kanneganti TD, Joosten LA, Netea MG (2012) The inflammasome puts obesity in the danger zone. Cell Metab 15:10–18
Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V (2011) Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J 30:4701–4711
O'Neill LA, Hardie DG (2013) Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature 493:346–355
Rodriguez-Prados JC, Traves PG, Cuenca J et al (2010) Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol 185:605–614
Giri S, Nath N, Smith B, Viollet B, Singh AK, Singh I (2004) 5-aminoimidazole-4-carboxamide-1-β-4-ribofuranoside inhibits proinflammatory response in glial cells: a possible role of AMP-activated protein kinase. J Neurosci 24:479–487
Jung CH, Jun CB, Ro SH et al (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20:1992–2003
Shanware NP, Bray K, Abraham RT (2013) The PI3K, metabolic, and autophagy networks: interactive partners in cellular health and disease. Annu Rev Pharmacol Toxicol 53:89–106
Ost A, Svensson K, Ruishalme I et al (2010) Attenuated mTOR signaling and enhanced autophagy in adipocytes from obese patients with type 2 diabetes. Mol Med 16:235–246
Kovsan J, Bluher M, Tarnovscki T et al (2011) Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab 96:E268–E277
Jansen HJ, van Essen P, Koenen T et al (2012) Autophagy activity is up-regulated in adipose tissue of obese individuals and modulates proinflammatory cytokine expression. Endocrinology 153:5866–5874
Nunez CE, Rodrigues VS, Gomes FS et al (2013) Defective regulation of adipose tissue autophagy in obesity. Int J Obes (2005) 37:1473–1480
Yoshizaki T, Kusunoki C, Kondo M et al (2012) Autophagy regulates inflammation in adipocytes. Biochem Biophys Res Commun 417:352–357
Brannmark C, Nyman E, Fagerholm S et al (2013) Insulin signaling in type 2 diabetes: experimental and modeling analyses reveal mechanisms of insulin resistance in human adipocytes. J Biol Chem 288:9867–9880
Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124:471–484
Cinti S, Mitchell G, Barbatelli G et al (2005) Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res 46:2347–2355
Miranda M, Escote X, Ceperuelo-Mallafre V et al (2005) Relation between human LPIN1, hypoxia and endoplasmic reticulum stress genes in subcutaneous and visceral adipose tissue. Int J Obes 34:679–686
Kaufman RJ (2002) Orchestrating the unfolded protein response in health and disease. J Clin Invest 110:1389–1398
Scherer T, Buettner C (2011) Yin and yang of hypothalamic insulin and leptin signaling in regulating white adipose tissue metabolism. Rev Endocr Metab Disord 12:235–243
Williams LM (2012) Hypothalamic dysfunction in obesity. Proc Nutr Soc 71:521–533
Zhang X, Zhang G, Zhang H, Karin M, Bai H, Cai D (2008) Hypothalamic IKKβ/NF-κB and ER stress link overnutrition to energy imbalance and obesity. Cell 135:61–73
Calegari VC, Torsoni AS, Vanzela EC et al (2011) Inflammation of the hypothalamus leads to defective pancreatic islet function. J Biol Chem 286:12870–12880
Meng Q, Cai D (2011) Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IκB kinase β (IKKβ)/NF-κB pathway. J Biol Chem 286:32324–32332
Leibowitz G, Kaiser N, Cerasi E (2009) Balancing needs and means: the dilemma of the beta-cell in the modern world. Diabetes Obes Metab 11(Suppl 4):1–9
Scheuner D, Kaufman RJ (2008) The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr Rev 29:317–333
Eizirik DL, Cardozo AK, Cnop M (2008) The role for endoplasmic reticulum stress in diabetes mellitus. Endocr Rev 29:42–61
Poitout V, Robertson RP (2008) Glucolipotoxicity: fuel excess and beta-cell dysfunction. Endocr Rev 29:351–366
Prentki M, Nolan CJ (2006) Islet beta cell failure in type 2 diabetes. J Clin Invest 116:1802–1812
Talchai C, Xuan S, Lin HV, Sussel L, Accili D (2012) Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell 150:1223–1234
Kroemer G, Marino G, Levine B (2010) Autophagy and the integrated stress response. Mol Cell 40:280–293
Mizushima N, Komatsu M (2011) Autophagy: renovation of cells and tissues. Cell 147:728–741
Orci L, Ravazzola M, Amherdt M et al (1984) Insulin, not C-peptide (proinsulin), is present in crinophagic bodies of the pancreatic B cell. J Cell Biol 98:222–228
Halban PA, Mutkoski R, Dodson G, Orci L (1987) Resistance of the insulin crystal to lysosomal proteases: implications for pancreatic B cell crinophagy. Diabetologia 30:348–353
Halban PA, Wollheim CB (1980) Intracellular degradation of insulin stores by rat pancreatic islets in vitro. An alternative pathway for homeostasis of pancreatic insulin content. J Biol Chem 255:6003–6006
Landstrom AH, Westman J, Borg LA (1988) Lysosomes and pancreatic islet function. Time course of insulin biosynthesis, insulin secretion, and lysosomal transformation after rapid changes in glucose concentration. Diabetes 37:309–316
Twig G, Elorza A, Molina AJ et al (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27:433–446
Kim I, Lemasters JJ (2011) Mitochondrial degradation by autophagy (mitophagy) in GFP-LC3 transgenic hepatocytes during nutrient deprivation. Am J Physiol Cell Physiol 300:C308–C317
Bommer G, Schafer HJ, Kloppel G (1976) Morphologic effects of diazoxide and diphenylhydantoin on insulin secretion and biosynthesis in B cells of mice. Virchows Arch A Pathol Anat Histol 371:227–241
Marsh BJ, Soden C, Alarcon C et al (2007) Regulated autophagy controls hormone content in secretory-deficient pancreatic endocrine beta-cells. Mol Endocrinol 21:2255–2269
Tomas A, Meda P, Regazzi R, Pessin JE, Halban PA (2008) Munc 18-1 and granuphilin collaborate during insulin granule exocytosis. Traffic 9:813–832
Rhodes CJ, Halban PA (1987) Newly synthesized proinsulin/insulin and stored insulin are released from pancreatic B cells predominantly via a regulated, rather than a constitutive, pathway. J Cell Biol 105:145–153
Uchizono Y, Alarcon C, Wicksteed BL, Marsh BJ, Rhodes CJ (2007) The balance between proinsulin biosynthesis and insulin secretion: where can imbalance lead? Diabetes Obes Metab 9(Suppl 2):56–66
Pearson GL, Mellett N, Chu KY et al (2014) Lysosomal acid lipase and lipophagy are constitutive negative regulators of glucose-stimulated insulin secretion from pancreatic beta cells. Diabetologia 57:129–139
Nair U, Jotwani A, Geng J et al (2011) SNARE proteins are required for macroautophagy. Cell 146:290–302
Jung HS, Chung KW, Won Kim J et al (2008) Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab 8:318–324
Ebato C, Uchida T, Arakawa M et al (2008) Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab 8:325–332
Quan W, Hur KY, Lim Y et al (2012) Autophagy deficiency in beta cells leads to compromised unfolded protein response and progression from obesity to diabetes in mice. Diabetologia 55:392–403
Kaniuk NA, Kiraly M, Bates H, Vranic M, Volchuk A, Brumell JH (2007) Ubiquitinated-protein aggregates form in pancreatic beta-cells during diabetes-induced oxidative stress and are regulated by autophagy. Diabetes 56:930–939
Masini M, Bugliani M, Lupi R et al (2009) Autophagy in human type 2 diabetes pancreatic beta cells. Diabetologia 52:1083–1086
Shen S, Kepp O, Kroemer G (2012) The end of autophagic cell death? Autophagy 8:1–3
Steneberg P, Bernardo L, Edfalk S et al (2013) The type 2 diabetes-associated gene ide is required for insulin secretion and suppression of alpha-synuclein levels in beta-cells. Diabetes 62:2004–2014
Las G, Serada SB, Wikstrom JD, Twig G, Shirihai OS (2011) Fatty acids suppress autophagic turnover in beta-cells. J Biol Chem 286:42534–42544
Rivera JF, Gurlo T, Daval M et al (2011) Human-IAPP disrupts the autophagy/lysosomal pathway in pancreatic beta-cells: protective role of p62-positive cytoplasmic inclusions. Cell Death Differ 18:415–426
Han D, Yang B, Olson LK et al (2010) Activation of autophagy through modulation of 5′-AMP-activated protein kinase protects pancreatic beta-cells from high glucose. Biochem J 425:541–551
Martino L, Masini M, Novelli M et al (2012) Palmitate activates autophagy in INS-1E beta-cells and in isolated rat and human pancreatic islets. PLoS One 7:e36188
Li S, Du L, Zhang L et al (2013) Cathepsin B contributes to Atg7-induced NLRP3 dependent pro-inflammatory response and aggravates lipotoxicity in rat insulinoma cell line. J Biol Chem 288:30094–30104
Saftig P, Beertsen W, Eskelinen EL (2008) LAMP-2: a control step for phagosome and autophagosome maturation. Autophagy 4:510–512
Tanaka Y, Guhde G, Suter A et al (2000) Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406:902–906
Fraenkel M, Ketzinel-Gilad M, Ariav Y et al (2008) mTOR inhibition by rapamycin prevents beta-cell adaptation to hyperglycemia and exacerbates the metabolic state in type 2 diabetes. Diabetes 57:945–957
Bachar E, Ariav Y, Ketzinel-Gilad M, Cerasi E, Kaiser N, Leibowitz G (2009) Glucose amplifies fatty acid-induced endoplasmic reticulum stress in pancreatic beta-cells via activation of mTORC1. PLoS One 4:e4954
Ogata M, Hino S, Saito A et al (2006) Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 26:9220–9231
Liu M, Hodish I, Rhodes CJ, Arvan P (2007) Proinsulin maturation, misfolding, and proteotoxicity. Proc Natl Acad Sci U S A 104:15841–15846
Hodish I, Liu M, Rajpal G et al (2010) Misfolded proinsulin affects bystander proinsulin in neonatal diabetes. J Biol Chem 285:685–694
Liu M, Haataja L, Wright J et al (2010) Mutant INS-gene induced diabetes of youth: proinsulin cysteine residues impose dominant-negative inhibition on wild-type proinsulin transport. PLoS One 5:e13333
Bartolome A, Guillen C, Benito M (2012) Autophagy plays a protective role in endoplasmic reticulum stress-mediated pancreatic beta cell death. Autophagy 8:1757–1768
Barlow AD, Nicholson ML, Herbert TP (2013) Evidence for rapamycin toxicity in pancreatic beta-cells and a review of the underlying molecular mechanisms. Diabetes 62:2674–2682
Tanemura M, Nagano H, Taniyama K, Kamiike W, Mori M, Doki Y (2012) Role of rapamycin-induced autophagy in pancreatic islets. Am J Transplant 12:1067
Fujimoto K, Hanson PT, Tran H et al (2009) Autophagy regulates pancreatic beta cell death in response to Pdx1 deficiency and nutrient deprivation. J Biol Chem 284:27664–27673
Wu J, Wu JJ, Yang LJ, Wei LX, Zou DJ (2013) Rosiglitazone protects against palmitate-induced pancreatic beta-cell death by activation of autophagy via 5′-AMP-activated protein kinase modulation. Endocrine 44:87–98
Koppaka S, Kehlenbrink S, Carey M et al (2013) Reduced adipose tissue macrophage content is associated with improved insulin sensitivity in thiazolidinedione-treated diabetic humans. Diabetes 62:1843–1854
Wasko MC, Hubert HB, Lingala VB et al (2007) Hydroxychloroquine and risk of diabetes in patients with rheumatoid arthritis. JAMA 298:187–193
Powrie JK, Smith GD, Shojaee-Moradie F, Sonksen PH, Jones RH (1991) Mode of action of chloroquine in patients with non-insulin-dependent diabetes mellitus. Am J Physiol 260:E897–E904
Shoji-Kawata S, Sumpter R, Leveno M et al (2013) Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494:201–206
Acknowledgements
We are grateful to E. Cerasi, Hadassah-Hebrew University Medical Center, for helpful discussions and to freelance graphic designer A. van der Kleij for superb technical assistance in the design of the figures. A. Rudich is Chair of the Fraida Foundation in Diabetes Research. G. Leibowitz is the Head of the Hebrew University Diabetes Research Center.
Funding
This work was supported by grants from Deutsche Forschungsgemeinschaft (DFG): SFB 1052/1: ‘Obesity mechanisms’ (project B02, to AR) and by the Israel Science Foundation (347/12) to GL. RS is supported by a VIDI-grant from the Netherlands Organization for Scientific Research (NWO).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
All authors were responsible for the conception and design of the manuscript, drafting the article and revising it critically for important intellectual content. All authors approved the version to be published.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Stienstra, R., Haim, Y., Riahi, Y. et al. Autophagy in adipose tissue and the beta cell: implications for obesity and diabetes. Diabetologia 57, 1505–1516 (2014). https://doi.org/10.1007/s00125-014-3255-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-014-3255-3