Abstract
Aims/hypothesis
The aim of this study was to examine human enteroviruses (HEVs) and other intestinal viruses derived from children who participated in the Babydiet intervention study and to analyse the findings according to the appearance of islet autoantibodies, dietary intervention, maternal type 1 diabetes and clinical symptoms.
Methods
In the Babydiet study the influence of first gluten exposure (6 or 12 months) on the development of islet autoimmunity was investigated in 150 children with increased genetic and familial risk for type 1 diabetes. Blood and stool samples were collected at 3 monthly intervals until the age of 3 years and yearly thereafter. Infections and clinical symptoms were recorded daily for the first year. In the present study, 339 stool samples collected from 104 children during the first year of life were analysed for HEVs and a certain proportion of the samples were analysed for other intestinal viruses.
Results
HEV was detected in 32 (9.4%) samples from 24 (23.1%) children. Altogether 13 serotypes were identified, with HEV-A species being the most common. Children with gastrointestinal symptoms had norovirus (3/11) and sapovirus (1/11) infections in addition to HEV (1/11). Of the 104 children, 22 developed islet autoantibodies. HEV infections were detected in 18% (4/22) and 24% (20/82) of islet-autoantibody-positive and -negative children, respectively (p = 0.5). The prevalence of HEV was similar in the gluten-exposed groups and in children from mothers with type 1 diabetes or from affected fathers and/or siblings (p = 1.0 and 0.6, respectively).
Conclusions/interpretation
No correlation was found between the presence of HEV in the first year of life and the development of islet autoantibodies. There was no association between HEV infections and dietary intervention, maternal diabetes or clinical symptoms.
Similar content being viewed by others
Introduction
The incidence of type 1 diabetes is increasing worldwide. While genetic factors contribute to this phenomenon, the role of environmental factors is clearly essential. Results from previous cross-sectional and prospective studies on type 1 diabetes patients and/or islet autoantibody seroconverting individuals have suggested that enterovirus infections are involved in the pathogenetic process [1–6]. Studies on pancreatic tissues from human autopsies and cultured human islets support the idea that during systemic enterovirus infections, the virus may reach pancreatic islets and cause direct beta cell damage [7–10]. Enterovirus genomes were detected in islets from postmortem pancreatic specimens of several type 1 diabetes patients but not in the corresponding samples from non-diabetic controls [7].
Human enteroviruses (HEVs) belong to the Enterovirus genus of the Picornaviridae family and include over 100 different serotypes divided into four species (HEV A–D) [11]. HEVs are small non-enveloped RNA viruses that cause infections that are very common, especially in early childhood, and peak in the HEV season extending from late summer to the end of the year [12, 13]. Enteroviruses are transmitted via the gastrointestinal or respiratory tract and replicate in the intestine and secondary target organs. HEV infections are mainly asymptomatic, but they are also associated with several different clinical manifestations, including: mild upper respiratory illness (common cold); flu-like symptoms; diarrhoea; hand, foot and mouth disease; herpangina; aseptic meningitis; encephalitis; and neonatal sepsis-like disease [13, 14].
In addition to HEVs, there are other RNA viruses that specifically infect the intestine and cause diarrhoea and are potentially associated with the pathogenetic processes prior to precipitation of autoimmune diseases. The potential role of rotavirus (RoV) infection in these processes has been reported several times [15–18]; cardiovirus infection can lead to systemic disease such as insulin-dependent diabetes in rodents [19–21]. In order to analyse potential connections between HEV infections and dietary factors in the process leading to clinical type 1 diabetes, we have investigated HEVs and other intestinal viruses derived from children with increased genetic and familial risk for type 1 diabetes participating in the Babydiet intervention study.
Methods
Babydiet study material
We investigated the presence of HEV infections in children participating in the Babydiet dietary intervention study (ClinicalTrials.gov no. NCT01115621) [22–25]. The Babydiet study determined whether delayed introduction of dietary gluten reduced the incidence of islet autoimmunity in high-risk first-degree relatives of patients with type 1 diabetes. The intervention in the first year of life consisted of delayed and controlled gluten exposure at the age of 6 months (control group) and 12 months (intervention group). Babydiet children were followed within the intervention study protocol until the age of 3 years, and 6 monthly thereafter as part of a natural history follow-up protocol. Babydiet study participants (n = 150) are offspring or siblings of patients with type 1 diabetes and have one of the following high-risk HLA genotypes: DRB1*03-DQA1*0501-DQB1*0201/DRB1*04-DQA1*0301-DQB1*0302; DRB1*04-DQA1*0301-DQB1*0302/DRB1*04-DQA1*0301-DQB1*0302; or DRB1*03-DQA1*0501-DQB1*0201/ DRB1*03-DQA1*0501-DQB1*0201.
Babydiet children were monitored intensively. Islet autoantibodies (insulin autoantibody [IAA], glutamic acid decarboxylase autoantibody [GADA], tyrosine phosphatase-like insulinoma antigen 2 antibody [IA-2A] and zinc transporter-8 antibody [ZnT8-A]) were measured at 3 monthly intervals. Details of infections, medication and the introduction of new food groups were recorded daily. Written informed consent was obtained from the parents. The study was approved by the ethics committee of the Ludwig-Maximilian-University, Munich, Germany (Ethikkommission der Medizinischen Fakultät der Ludwig-Maximilians Universität No. 329/00).
The Babydiet protocol scheduled the collection of stool samples every 3 months from birth to the age of 3 years. For this HEV study, all stool samples available from the first year of life (n = 339 faecal specimens) were included (electronic supplementary material [ESM] Fig. 1). The specimens were collected from 104 children at 2–12 months of age between December 2000 and June 2007 (ESM Fig. 2). The remaining 38 children were not compliant with stool sample collection and eight dropped out of the study before reaching 12 months of age and were also not included in the analyses. The number of specimens per child varied from one to five. Eleven of 339 faecal specimens were collected from children with gastrointestinal symptoms. From these samples the presence of other enteric viruses (norovirus [NoV] genogroup I [GI] and genogroup II [GII], astrovirus [AsV], RoV and sapovirus [SaV]) was also investigated. In addition, all stool samples from 12-month-old children were screened for the human cardiovirus, Saffold virus (SAFV).
Collection of infant diet, infection and medication data
All families received a Babydiet book in which to record food, infections, illnesses and medications for the Babydiet children daily between the ages of 3 and 18 months. In particular, the introduction of new food groups and data on breastfeeding (yes, no) and the duration of ‘full’ breastfeeding (weeks) and ‘any’ breastfeeding were recorded by the parents. Breastfeeding was defined according to WHO criteria [26] as full breastfeeding if the infant received breast milk with or without supplementation with water or water-based drinks, vitamins and medicines, but without formula or other milk or solids, or as any breastfeeding if the infant received breast milk irrespective of any other types of food including full breastfeeding. Detailed data regarding fever (defined as temperature >38°C), gastrointestinal infections and treatment with systemic antibiotics were taken from the infectious disease and medication records written daily by the child’s parents. The parents documented the number of days with fever, the number of days with infectious symptoms, the type of symptom, and the name and days of administered pharmaceutical agents or their active ingredient.
Processing of stool samples and RNA isolation
A 10% (wt/vol.) stool extract was prepared by shaking with glass beads the stool specimen with PBS with benzylpenicillin (100 U/ml), streptomycin sulphate (100 μg/ml), gentamicin sulphate (50 μg/ml) and chloroform. After centrifugation (1,118 g for 20 min), stool extract supernatant fraction was collected. Viral RNA was isolated from 100 μl of stool extract using E.Z.N.A. Total RNA kit (Omega Bio-Tek, Doraville, GA, USA).
Gene amplification
Extracted RNAs were analysed for HEV, NoV, AsV, RoV and SaV using validated real-time methods based on TaqMan chemistry. References for primers used in different PCRs were: Le Cann et al. for AsV [27]; Kilpatrick et al. for HEV [28]; Maunula et al. for NoV GI [29]; Loisy et al. for NoV GII [30]; Gutierrez-Aguirre et al. for RoV [31]; and Svraka et al. for SaV [32]. The cut-off threshold value was set at 10% of the strongest positive control. Curves rising above the threshold were considered positive. RNA samples were screened for SAFV with an in-house real-time RT-PCR method using the EXPRESS One-Step SYBR GreenER Kit (Invitrogen Corporation, Carlsbad, CA, USA) and primers described in Chiu et al. [33]. All real-time assays were carried out using an Mx3005P analyser (Stratagene, La Jolla, CA, USA).
HEV isolation
Virus isolation was carried out in monolayer cultures of cells of the human rhabdomyosarcoma cell line (RD). Eagle’s minimal essential medium (containing 5% [vol./vol.] fetal bovine serum, 20 mmol/l HEPES, pH 7.4, 20 mmol/l MgCl2, benzylpenicillin [100 U/ml], streptomycin sulphate [100 μg/ml], gentamicin sulphate [50 μg/ml] and nystatin [50 U/ml]) was used as a growth medium. Cells were inoculated with stool extract (200 μl) and the cultures were incubated at 36°C in 5% CO2 for 7 days and monitored microscopically for virus-induced cytopathic effect (CPE). Viral RNA was isolated from 100 μl of virus-positive cell culture suspension as above. Protocols for HEV detection, isolation and identification are accredited according to the international standard EN ISO/IEC 17025:2005 by the Finnish Accreditation Service FINAS.
HEV genetic typing and sequence and phylogenetic analyses
For typing the virus isolates obtained from virus-positive cell culture suspensions, RT-PCR for a partial 5′ part of the capsid protein VP1 coding region was performed as described by Oberste et al. [34] using primers 222-reverse and 292-forward. If the virus isolate was not successfully amplified, RT-PCR for a partial 3′ part of the VP1 region was performed with the primers 012- and 040-forward and 011-reverse [35]. RT-PCR products were visualised after electrophoresis on an ethidium-bromide-stained 2% (wt/vol.) agarose gel and purified prior to sequencing with an Elchrom Gel Purification kit (Elchrom Scientific, Cham, Switzerland). For typing of the isolates infecting cell cultures weakly or not at all, semi-nested (sn) RT-PCR for a partial VP1 region was performed according to Nix et al. [36] using RNA obtained from the stool extract as a starting material. Primers AN32-35 for cDNA, SO224-forward and SO222-reverse for the first PCR and AN88-reverse and AN89-forward for the second PCR were used. The products of sn-RT-PCR were visualised after electrophoresis on an ethidium-bromide-stained 1.2% (wt/vol.) agarose gel and purified prior to sequencing with QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). The purified products were sequenced at the Molecular Medicine Sequencing Laboratory (Biomedicum, Helsinki, Finland), with the same primer pairs as used in RT-PCR and sn-RT-PCR. Nucleotide sequence data were analysed with ContigExpress (Vector NTI Advance 10.1; Invitrogen). Nucleotide sequences were compared with sequences in GenBank using the BLAST nucleotide–nucleotide search tool [37].
Multiple sequence alignments were made using the Clustal W algorithm embedded in Molecular Evolutionary Genetics Analysis (MEGA) version 4.0 [38]. Phylogenetic trees were produced and visualised with MEGA version 4.0 using the neighbour-joining method [39]. Evolutionary distances were computed using the Tamura–Nei model [40]. Bootstrap analysis was performed using 1,000 replicates [41].
Using the identified sequence data of the designated Babydiet isolates as query sequences, other available sequences of the corresponding serotypes were obtained from GenBank and from the local HEV database. The sequences generated in this study have been submitted to GenBank under accession numbers JN034197–JN034251.
Statistical analysis
The prevalence of HEV infections was determined and compared between children developing islet autoantibodies and children who remained islet-autoantibody-negative using the χ 2 test. The relationship of HEV infections to proband, diet and symptoms was also tested with the χ 2 test. Multivariate analyses of the influence of HEV infection, dietary intervention, proband and breastfeeding on the appearance of islet autoantibodies were performed using the general linear model. All p values were two-tailed. Statistical analyses were performed using the Statistical Package for Social Science (PASW Statistics 18.0; SPSS, Chicago, IL, USA).
Results
HEV screening
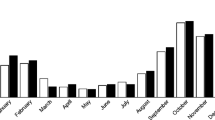
Altogether, 32 of 339 (9.4%) stool samples were positive for HEV in real-time RT-PCR, and 24 of the 104 (23.1%) children examined had one or more HEV infections during the first year of life (Fig. 1). In addition, five stool samples had fluorescence values in real-time RT-PCR that were increased but remained under the threshold limit described above. These samples were designated ‘borderline cases’.
a Prevalence of HEV infections in stool samples and children within the first year of life. b Prevalence of HEV infections in stool samples and children within the first year of life relative to islet autoantibody positivity. The p values for comparison of children and stool samples according to autoantibody positivity are 0.5 and 0.4, respectively
HEV isolation and identification
In order to identify virus strains and serotypes responsible for infections, HEVs were isolated with a traditional viral culturing method using the RD cell line. To confirm the result of the HEV-specific real-time RT-PCR all the samples were cultured. Altogether 62 specimens were detected that caused cell destruction with minor to total CPE. This number comprises 27 positive and 31 negative specimens in HEV real-time RT-PCR as well as four borderline cases. Samples from five real-time RT-PCR-positive and one borderline case did not have CPE in the cell line used. Samples positive in viral culturing with total CPE were identified to a serotype level by partial sequencing of the capsid protein VP1-coding region using virus-positive cell culture suspension as a starting material; samples negative or weakly positive in viral culturing but positive in real-time RT-PCR were identified using the more sensitive sn-RT-PCR and partial sequencing of the VP1 coding region using stool extract as a starting material. A serotype was identified from 30 and one of the real-time RT-PCR-positive and borderline case samples, respectively (31/37; 83.8%). Despite the CPE-like cell destruction, the real-time RT-PCR-negative samples remained negative in VP1 RT-PCR, confirming the result of the HEV-specific real-time RT-PCR. In total, identified HEV was detected in 9.1% (31/339) of the samples. One or more samples were HEV-positive with a confirmed serotype in 21.2% (22/104) of the children.
Identified HEV species and serotypes
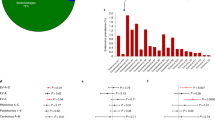
Based on partial VP1 sequences, a total of 13 different serotypes were detected (Fig. 2a). Serotypes belonging to HEV-A and HEV-B species covered 64.5% and 35.5% of the findings, respectively, with an overall prevalence of 5.9% (20/339) and 3.2% (11/339), respectively. Neither HEV-C nor HEV-D group viruses were detected. The most common serotypes were coxsackievirus (CV)-A4, CV-A2, CV-A10 and echovirus (E)-25, accounting for 19.4%, 16.1%, 16.1% and 12.9% of the findings, respectively.
Seasonal variation of HEV infections
The majority of the HEV infections occurred during the enterovirus season (80.6% between July and December; data not shown). CV-A2 was detected during a year-long interval from July 2004 to July 2005, whereas CV-A4, CV-A10 and E-25 were detected during longer time periods (Fig. 2b).
Age-specific frequency of HEV infection
The number (11) and the frequency (22.4%) of HEV infections were highest at the age of 12 months (Fig. 3). The HEV findings of the 12-month-olds included 45.5% and 54.5% of HEV-A and HEV-B isolates, respectively (data not shown). The children younger than 12 months had mainly HEV-A isolates (75% of the findings; Fig. 2a).
HEV infection in relation to islet autoantibody seroconversion
Of the 104 children included in the HEV study, 22 developed islet autoantibodies (ESM Fig. 1, ESM Table 1). Four (18%) of these children had stool samples infected with HEV during the first year of life prior to the date of seroconversion. In contrast, 20 (24%) children who remained islet-autoantibody negative had HEV infections during the first year of life (p = 0.5; Fig. 1). Of the four children with HEV infections prior to seroconversion, one had two HEV infections with distinct identified causative agents (CV-A6 and CV-B2 in samples 92 and 94, respectively) and three had one infection (CV-A2 in sample 177, E-9 in sample 79 and E-25 in sample 28). A multivariate analysis including HEV infections, dietary intervention, proband and breastfeeding was performed to determine the association of these variables with the appearance of islet autoantibodies. None of these variables was significantly associated with the development of islet autoantibodies, nor with the rate of HEV infections.
Relation of HEV infection to proband, diet and symptoms
The prevalence of HEV infections was similar in children from mothers with type 1 diabetes (25%) compared with children with affected fathers and/or siblings (21%, p = 0.6). The prevalence of HEV infections was also comparable in children who were introduced to gluten at age 6 months (23%) or 12 months (23%, p = 1.0). Breastfeeding duration was 6.5 months in children with HEV infections compared with 6.4 months in children without HEV infections. Seven of the 24 children with HEV infections were breastfed at the time of HEV infection. There was no association of HEV infection and the presence of clinical symptoms. Information on corresponding clinical symptoms was available for 266 stool samples. Respiratory, gastrointestinal and other symptoms (such as otitis media) around the time of stool sampling were reported in 44%, 4% and 8% of HEV-positive stool samples and in 40.2%, 4.1% and 4.6% of HEV-negative stool samples.
Genetic characterisation of the HEV findings
The partial VP1 sequences of the frequently detected serotypes were compared with sequences in GenBank and phylogenetic trees were constructed in order to characterise genetic relationships. The clustering of the most common strains of this study (CV-A2, -A4, -A10 and E-25) indicates them to be similar to strains circulating in the community in general (data not shown except for CV-A2 and E-25 in ESM Fig. 3). The phylogenetic trees of the strains derived from children who seroconverted to become autoantibody positive during the Babydiet intervention study (shown in ESM Table 1) as well as other strains of the given type identified in this study are presented in ESM Fig. 3. Similarly, the autoantibody-positive strains clustered among the strains circulating in general. The CV-A2 phylogeny suggests wide geographic circulation of distinct genotypes. The phylogenetic analysis of CV-A6 sequences revealed three major lineages. The virus strain isolated from sample 92 grouped together with the strains isolated from Scandinavia. However, both CV-B2 and E-25 formed multiple temporally and geographically co-circulating lineages. The CV-B2 strains isolated from samples 94 and 210 clustered individually with Finnish and other European strains. The E-25 strains affiliated to clusters consisting of concomitantly circulating strains from Europe and Australia. E-9 strains formed a tree-trunk-like phylogeny with several inherently evolving sublineages. Both the diabetogenic E-9-DM strain isolated from a 6-week-old child at the clinical onset of type 1 diabetes [42] and the virus strain isolated from sample 79 are members of a large cluster with worldwide circulation. Strain 79 showed a close relationship to strains circulating in Western Europe and Australia at the time of isolation.
Other intestinal RNA viruses detected
Eleven children had gastrointestinal symptoms during the first year of life (stomach pain, diarrhoea, constipation, nausea or vomiting) including four children with islet autoantibodies (children 7, 9, 12 and 19; ESM Table 1). Altogether 11 specimens collected from children with gastrointestinal symptoms were analysed, in addition to HEV, for the presence of other intestinal RNA viruses (NoV, AsV, RoV and SaV). Virus-specific real-time RT-PCR methods revealed that five of the studied faecal specimens were positive for enteric viruses (one for HEV, three for NoV and one for SaV). Two of the islet-autoantibody-positive children (numbers 7 and 9) had NoV infections and one (number 19) had an SaV infection. All the specimens derived from 12-month-olds, the age group of German children presumed to be just starting in day care, were screened for SAFV. The results of the in-house real-time RT-PCR method showed that all 49 stool samples analysed were negative for SAFV.
Discussion
In the pathogenesis of type 1 diabetes, intestinal events and factors entering the body via the intestine (e.g. infectious agents and nutritious substances) are considered to be of great importance. In order to demonstrate a potential connection of HEV infections and dietary factors in the process leading to clinical type 1 diabetes we have analysed stool samples collected from children with a higher risk for the development of type 1 diabetes for the presence of HEV infections with a validated real-time RT-PCR method. This material is unique for characterisation of early events in the process potentially leading to type 1 diabetes. The results of HEV detection were confirmed with a cell culture isolation method, the gold standard for HEV detection. While about 10% of the specimens tested positive for HEV, in accordance with other recent studies [43], and several distinct serotypes of HEV were identified, no correlation was observed between HEV positivity in German children and the development of islet autoantibodies compared with HEV-negative children.
It has been speculated that autoantibody-positive patients may have been infected with specific diabetogenic virus types. In this study five strains were isolated from autoantibody-positive children during the first year of life prior to the date of seroconversion. These strains were phylogenetically compared with strains isolated from autoantibody-negative children in this study as well as other reference strains. The strains isolated from children who tested positive for autoantibodies did not form phylogenetically distinct lineages, but clustered together with other circulating strains of respective types.
Viruses associated with the development of type 1 diabetes are likely to be the same viruses frequently circulating in communities. Most of our HEV findings belonged to HEV-A species, often considered rare and hard to detect with traditional virus isolation assays. Previously, HEV-A strains have been detected with a sensitive PCR method [44]. In this study, the majority of the HEV-A strains detected were also cell-culture positive. One of the observed HEV-A strains was CV-A6, which is known to have circulated in Europe [45] and to have caused a large outbreak in Finland subsequent to sampling the German children of the Babydiet study [46, 47]. These findings suggest that the discovery of HEV serotype pattern depends on the epidemiological situation and geographical area sampled. It is known that HEV-B types frequently circulate in communities [48] and are therefore readily isolated in surveillance studies that focus on diabetes-prone children. However, there is no reason to assume that viruses of other HEV species are less capable of infecting pancreatic beta cells or having a potential role in the process leading to type 1 diabetes [10, 49].
The potential diabetogenic properties of enteroviruses may not be serotype specific, but large variations exist among virus strains of the same serotype in terms of capability of causing functional impairment and damage in pancreatic islet beta cells [10, 42]. The quasi-species nature of enteroviruses may mean that beta cell tropic and potentially diabetogenic variants can sporadically emerge during systemic enterovirus infection. Moreover, the potential diabetogenic properties are possibly defined partly by viral-strain-specific characteristics other than those defining or associated with serotype characteristics. These characteristics remain unnoticed in the molecular epidemiological analyses based on genomic capsid-coding sequences used for genetic typing. The pathogenetic properties of an enterovirus strain are partly independent of the serotype or genetic sublineage of a given isolate, and may vary greatly among strains within a serotype. For example, certain strains of E-11 cause haemorrhagic uveitis [50], while their close genetic relatives are abundant in environmental sewage samples, reflecting wide circulation in a community without a drastic clinical outbreak [51].
According to our hypothesis other intestinal infections may also be of importance in the process leading to type 1 diabetes. We explored the presence of other enteric viruses in the Babydiet material. Enteric viruses such as AsVs, RoVs, SaVs and NoVs are a major cause of acute gastroenteritis, one of the most common diseases in humans worldwide. RoVs, AsVs and SaVs cause infections primarily in children, while NoVs cause infections in all age groups. SaVs and NoVs were indeed detected in the Babydiet material, but because of the limited number of specimens studied here, the correlation of these agents and development of type 1 diabetes remains to be further elucidated. No SAFVs were detected among the 12-month-old children, presumably just starting in day care, despite the observed frequency [52] of this recently identified pathogen. However, the number of specimens studied was small, suggesting that no drastic conclusions can be drawn.
The association of HEVs with the development of autoimmunity or type 1 diabetes has been shown in several studies, including a recent meta-analysis [6]. However, there are also studies reporting a negative association [43]. The contradictory results observed for the association of enteroviruses and type 1 diabetes can potentially be explained by differences in the epidemic situation of enteroviral infectivity at the time of sample collection as well as by methods for the enterovirus detection. As enteroviruses are easily transmitted and circulate frequently, it is possible that the observed positive link to the future development of type 1 diabetes is only revealed at a favourable epidemic situation (i.e. at a time of outbreak-like enterovirus circulation). Genetic factors will also affect results [6].
As a conclusion, the results of the Babydiet study show that HEV infection in the first year of life was not associated with dietary intervention, maternal diabetes, autoantibody development or reported clinical symptoms.
Abbreviations
- AsV:
-
Astrovirus
- CPE:
-
Cytopathic effect
- CV:
-
Coxsackievirus
- E:
-
Echovirus
- GI:
-
Genogroup I
- GII:
-
Genogroup II
- GADA:
-
Glutamic acid decarboxylase autoantibody
- HEV:
-
Human enterovirus
- IA-2A:
-
Tyrosine phosphatase-like insulinoma antigen 2 antibody
- IAA:
-
Insulin autoantibody
- NoV:
-
Norovirus
- RD:
-
Human rhabdomyosarcoma cell line
- RoV:
-
Rotavirus
- SAFV:
-
Saffold virus
- SaV:
-
Sapovirus
- Sn:
-
Semi-nested
- ZnT8-A:
-
Zinc transporter-8 antibody
References
Clements GB, Galbraith DN, Taylor KW (1995) Coxsackie B virus infection and onset of childhood diabetes. Lancet 346:221–223
Hyoty H, Hiltunen M, Knip M et al (1995) A prospective study of the role of coxsackie B and other enterovirus infections in the pathogenesis of IDDM. Childhood Diabetes in Finland (DiMe) Study Group. Diabetes 44:652–657
Andreoletti L, Hober D, Hober-Vandenberghe C et al (1997) Detection of coxsackie B virus RNA sequences in whole blood samples from adult patients at the onset of type I diabetes mellitus. J Med Virol 52:121–127
Roivainen M, Knip M, Hyoty H et al (1998) Several different enterovirus serotypes can be associated with prediabetic autoimmune episodes and onset of overt IDDM. Childhood Diabetes in Finland (DiMe) Study Group. J Med Virol 56:74–78
Chehadeh W, Weill J, Vantyghem MC et al (2000) Increased level of interferon-alpha in blood of patients with insulin-dependent diabetes mellitus: relationship with coxsackievirus B infection. J Infect Dis 181:1929–1939
Yeung WC, Rawlinson WD, Craig ME (2011) Enterovirus infection and type 1 diabetes mellitus: systematic review and meta-analysis of observational molecular studies. BMJ 342:d35
Ylipaasto P, Klingel K, Lindberg AM et al (2004) Enterovirus infection in human pancreatic islet cells, islet tropism in vivo and receptor involvement in cultured islet beta cells. Diabetologia 47:225–239
Dotta F, Censini S, van Halteren AG et al (2007) Coxsackie B4 virus infection of beta cells and natural killer cell insulitis in recent-onset type 1 diabetic patients. Proc Natl Acad Sci USA 104:5115–5120
Richardson SJ, Willcox A, Bone AJ, Foulis AK, Morgan NG (2009) The prevalence of enteroviral capsid protein vp1 immunostaining in pancreatic islets in human type 1 diabetes. Diabetologia 52:1143–1151
Roivainen M, Ylipaasto P, Savolainen C, Galama J, Hovi T, Otonkoski T (2002) Functional impairment and killing of human beta cells by enteroviruses: the capacity is shared by a wide range of serotypes, but the extent is a characteristic of individual virus strains. Diabetologia 45:693–702
International Committee on Taxonomy of Viruses (ICTV) Virus taxonomy: 2009 release. Available from www.ictvonline.org/virusTaxonomy.asp?version=2009. Accessed 1 February 2011
Hovi T, Stenvik M, Rosenlew M (1996) Relative abundance of enterovirus serotypes in sewage differs from that in patients: clinical and epidemiological implications. Epidemiol Infect 116:91–97
Pallansch MA, Roos RP (2001) Enteroviruses: polioviruses, coxsackieviruses, echoviruses and newer enteroviruses. In: Knipe DM, Howley PM, Griffin DE et al (eds) Field’s virology. Lippincot, Williams and Wilkins, Philadelphia, pp 723–775
Zaoutis T, Klein JD (1998) Enterovirus infections. Pediatr Rev 19:183–191
Nigro G (1991) Pancreatitis with hypoglycemia-associated convulsions following rotavirus gastroenteritis. J Pediatr Gastroenterol Nutr 12:280–282
Honeyman MC, Coulson BS, Stone NL et al (2000) Association between rotavirus infection and pancreatic islet autoimmunity in children at risk of developing type 1 diabetes. Diabetes 49:1319–1324
Coulson BS, Witterick PD, Tan Y et al (2002) Growth of rotaviruses in primary pancreatic cells. J Virol 76:9537–9544
Honeyman MC, Stone NL, Falk BA, Nepom G, Harrison LC (2010) Evidence for molecular mimicry between human T cell epitopes in rotavirus and pancreatic islet autoantigens. J Immunol 184:2204–2210
Craighead JE, McLane MF (1968) Diabetes mellitus: induction in mice by encephalomyocarditis virus. Science 162:913–914
Boucher DW, Notkins AL (1973) Virus-induced diabetes mellitus. I. Hyperglycemia and hypoinsulinemia in mice infected with encephalomyocarditis virus. J Exp Med 137:1226–1239
Notkins AL (1979) The causes of diabetes. Sci Am 241:62–73
Schmid S, Buuck D, Knopff A, Bonifacio E, Ziegler AG (2004) Babydiet, a feasibility study to prevent the appearance of islet autoantibodies in relatives of patients with type 1 diabetes by delaying exposure to gluten. Diabetologia 47:1130–1131
Pfluger M, Winkler C, Hummel S, Ziegler AG (2010) Early infant diet in children at high risk for type 1 diabetes. Horm Metab Res 42:143–148
Boerschmann H, Pfluger M, Henneberger L, Ziegler AG, Hummel S (2010) Prevalence and predictors of overweight and insulin resistance in offspring of mothers with gestational diabetes mellitus. Diabetes Care 33:1845–1849
Hummel S, Pfluger M, Hummel M, Bonifacio E, Ziegler AG (2011) Primary dietary intervention study to reduce the risk of islet autoimmunity in children at increased risk for type 1 diabetes: the Babydiet study. Diabetes Care 34:1301–1305
WHO (2001) The optimal duration of exclusive breastfeeding: results of a WHO systematic review. Indian Pediatr 38:565–567
Le Cann P, Ranarijaona S, Monpoeho S, Le Guyader F, Ferré V (2004) Quantification of human astroviruses in sewage using real-time RT-PCR. Res Microbiol 155:11–15
Kilpatrick DR, Yang CF, Ching K et al (2009) Rapid group-, serotype-, and vaccine strain-specific identification of poliovirus isolates by real-time reverse transcription-PCR using degenerate primers and probes containing deoxyinosine residues. J Clin Microbiol 47:1939–1941
Maunula L, Klemola P, Kauppinen A et al (2009) Enteric viruses in a large waterborne outbreak of acute gastroenteritis in Finland. Food Environ Virol 1:31–36
Loisy F, Atmar RL, Guillon P, Le Cann P, Pommepuy M, Le Guyader FS (2005) Real-time RT-PCR for norovirus screening in shellfish. J Virol Methods 123:1–7
Gutierrez-Aguirre I, Steyer A, Boben J, Gruden K, Poljsak-Prijatelj M, Ravnikar M (2008) Sensitive detection of multiple rotavirus genotypes with a single reverse transcription-real-time quantitative PCR assay. J Clin Microbiol 46:2547–2554
Svraka S, van der Veer B, Duizer E, Dekkers J, Koopmans M, Vennema H (2009) Novel approach for detection of enteric viruses to enable syndrome surveillance of acute viral gastroenteritis. J Clin Microbiol 47:1674–1679
Chiu CY, Greninger AL, Kanada K et al (2008) Identification of cardioviruses related to Theiler’s murine encephalomyelitis virus in human infections. Proc Natl Acad Sci USA 105:14124–14129
Oberste MS, Nix WA, Maher K, Pallansch MA (2003) Improved molecular identification of enteroviruses by RT-PCR and amplicon sequencing. J Clin Virol 26:375–377
Oberste MS, Maher K, Kilpatrick DR, Pallansch MA (1999) Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol 73:1941–1948
Nix WA, Oberste MS, Pallansch MA (2006) Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol 44:2698–2704
Basic Local Alignment Search Tool (BLAST). Available from www.ncbi.nlm.nih.gov/BLAST/Blast.cgi. Accessed 19 May 2010–23 March 2011
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Paananen A, Ylipaasto P, Rieder E, Hovi T, Galama J, Roivainen M (2003) Molecular and biological analysis of echovirus 9 strain isolated from a diabetic child. J Med Virol 69:529–537
Tapia G, Cinek O, Rasmussen T et al (2011) Human enterovirus RNA in monthly fecal samples and islet autoimmunity in Norwegian children with high genetic risk for type 1 diabetes: the MIDIA study. Diabetes Care 34:151–155
Witso E, Palacios G, Cinek O et al (2006) High prevalence of human enterovirus a infections in natural circulation of human enteroviruses. J Clin Microbiol 44:4095–4100
Davia JL, Bel PH, Ninet VZ et al (2011) Onychomadesis outbreak in Valencia, Spain associated with hand, foot, and mouth disease caused by enteroviruses. Pediatr Dermatol 28:1–5
Blomqvist S, Klemola P, Kaijalainen S et al (2010) Co-circulation of coxsackieviruses A6 and A10 in hand, foot and mouth disease outbreak in Finland. J Clin Virol 48:49–54
Osterback R, Vuorinen T, Linna M, Susi P, Hyypia T, Waris M (2009) Coxsackievirus A6 and hand, foot, and mouth disease, Finland. Emerg Infect Dis 15:1485–1488
Klemola P, Kaijalainen S, Ylipaasto P, Roivainen M (2008) Diabetogenic effects of the most prevalent enteroviruses in Finnish sewage. Ann NY Acad Sci 1150:210–212
Smura T, Ylipaasto P, Klemola P et al (2010) Cellular tropism of human enterovirus D species serotypes EV-94, EV-70, and EV-68 in vitro: implications for pathogenesis. J Med Virol 82:1940–1949
Lukashev AN, Lashkevich VA, Koroleva GA et al (2003) Molecular epidemiology of enteroviruses causing uveitis and multisystem hemorrhagic disease of infants. Virology 307:45–53
Savolainen-Kopra C, Al-Hello H, Paananen A et al (2009) Molecular epidemiology and dual serotype specificity detection of echovirus 11 strains in Finland. Virus Res 139:32–38
Zoll J, Erkens Hulshof S, Lanke K et al (2009) Saffold virus, a human Theiler’s-like cardiovirus, is ubiquitous and causes infection early in life. PLoS Pathog 5:e1000416
Acknowledgements
The study was supported by grants from Deutsche Forschungsgemeinschaft (DFG ZI-310/14-1 to-4), the foundation ‘Children With Type 1 Diabetes’ (Stiftung Das Zuckerkranke Kind), the German Association for Coeliac Disease (Deutsche Zöliakiegesellschaft), the Institute Danone Nutrition for Health, the German Association for Clinical Nutrition, the German Competence Net for Diabetes (grant 01GI0805-07), the European Union (EP7-HEALTH-2007, DIAPREPP N202013), Academy of Finland, and the Juvenile Diabetes Research Foundation (USA). The authors thank A. Knopff (Forschergruppe Diabetes e.V. at Helmholtz Center Munich, Neuherberg, Germany), U. Mollenhauer (Forschergruppe Diabetes e.V. at Helmholtz Center Munich, Neuherberg, Germany), M. Zwilling (Forschergruppe Diabetes e.V. at Helmholtz Center Munich, Neuherberg, Germany) and A. Paananen (Intestinal Viruses Unit, National Institute for Health and Welfare, Helsinki, Finland) for expert technical assistance.
Contribution statement
MLST, PK, CSK, TS, SK, KN and ON analysed clinical samples for different viruses, analysed data, performed phylogenetic analyses, interpreted the data and wrote the manuscript. MR, as the principal investigator of the viral analyses, designed this part of the study and was involved in the interpretation of the results and writing of the manuscript and critically reviewed the manuscript for intellectual content. MP researched data, undertook statistical analysis, interpreted the results and reviewed and edited the manuscript. SH coordinated the Babydiet study, assisted in obtaining the data, undertook statistical analyses and critically reviewed the manuscript. AZ is the principal investigator, designed the Babydiet study and HEV substudy, was involved in the interpretation of the results and writing of the manuscript, and critically reviewed the manuscript for intellectual content. The final version of the manuscript was approved by all the authors.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
M. Roivainen and A.-G. Ziegler contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM Table 1
(PDF 34 kb)
ESM Fig. 1
Flow diagram of the BABYDIET study population and the samples included in the HEV study. (PDF 18 kb)
ESM Fig. 2
Samples included in the HEV study according to the sampling month (from December 2000 to June 2007). The ages of children during sampling are shown with colours. (PDF 10 kb)
ESM Fig. 3
Phylogenetic trees constructed from the partial 5’-end VP1 nucleotide sequences (220–317 nucleotides) using the neighbour-joining method with MEGA version 4.0. The bootstrap analysis was performed using 1000 replicates and the evolutionary distances were computed using the Tamura-Nei model. Sequences generated in the present study are labelled with black spheres (isolates from autoantibody positive children) and black triangles (autoantibody negative). The isolates derived from GenBank and the local HEV database were included in the analyses. In case of numerous findings, the sequences retrieved from GenBank were delimited in order to represent all variable findings within given country and isolation year. GenBank accession number as well as the isolation country and year, if available, are shown for the sequences retrieved from GenBank. (PDF 23 kb)
Rights and permissions
About this article
Cite this article
Simonen-Tikka, ML., Pflueger, M., Klemola, P. et al. Human enterovirus infections in children at increased risk for type 1 diabetes: the Babydiet study. Diabetologia 54, 2995–3002 (2011). https://doi.org/10.1007/s00125-011-2305-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-011-2305-3