Abstract
Aims/hypothesis
The destruction of pancreatic beta cells leading to type 1 diabetes in humans is thought to occur mainly through apoptosis and necrosis induced by activated macrophages and T cells, and in which secreted cytokines play a significant role. The transcription factor nuclear factor kappa-B (NF-κB) plays an important role in mediating the apoptotic action of cytokines in beta cells. We therefore sought to determine the changes in expression of genes modulated by NF-κB in human islets exposed to a combination of IL1β, TNF-α and IFN-γ.
Methods
Microarray and gene set enrichment analysis were performed to investigate the global response of gene expression and pathways modulated in cultured human islets exposed to cytokines. Validation of a panel of NF-κB-regulated genes was performed by quantitative RT-PCR. The mechanism of induction of BIRC3 by cytokines was examined by transient transfection of BIRC3 promoter constructs linked to a luciferase gene in MIN6 cells, a mouse beta cell line.
Results
Enrichment of several metabolic and signalling pathways was observed in cytokine-treated human islets. In addition to the upregulation of known pro-apoptotic genes, a number of anti-apoptotic genes including BIRC3, BCL2A1, TNFAIP3, CFLAR and TRAF1 were induced by cytokines through NF-κB. Significant synergy between the cytokines was observed in NF-κB-mediated induction of the promoter of BIRC3 in MIN6 cells.
Conclusions/interpretation
These findings suggest that, via NF-κB activation, cytokines induce a concurrent anti-apoptotic pathway that may be critical for preserving islet integrity and viability during the progression of insulitis in type 1 diabetes.
Similar content being viewed by others
Introduction
Type 1 diabetes is an autoimmune disorder characterised by immune cell-mediated destruction of the beta cells in the islets of Langerhans in pancreas [1, 2]. In female NOD mice, a model of type 1 diabetes, autoimmunity is evident at approximately 3 to 4 weeks of age as infiltration of the perivascular ducts and peri-islet regions of the pancreas, initially by macrophages and dendritic cells, and subsequently by B and T lymphocytes [3]. Activated macrophages and T cells secrete soluble mediators like cytokines, including IL1β, TNF-α and IFN-γ, chemokines, nitric oxide and oxygen free radicals, which impair beta cell function and subsequently cause beta cell destruction, with overt disease typically occurring by 4 to 6 months of age [4, 5]. Although, the natural history of human type 1 diabetes is temporally more variable and is accompanied by less insulitis, the progression of autoimmune disease is similar to that in NOD mice [6]. The mode of beta cell death is considered to be primarily through apoptosis in rodent and human islets [7, 8]. Under in vitro conditions, acute exposure to IL1-β alone or in combination with TNF-α and/or IFN-γ induces severe beta cell dysfunction and death by apoptotic and necrotic processes in rodent islets [9, 10].
Cytokines are known to activate nuclear factor kappa-B (NF-κB) pathway leading to changes in the expression of genes involved in apoptosis as well as in cell survival [11]. The transcription factor NF-κB consists of multiple subunits, including v-rel reticuloendotheliosis viral oncogene homologue A (RELA)/p65, v-rel reticuloendotheliosis viral oncogene homologue (avian) (cREL), bifunctional antitoxin/transcriptional repressor RelB (RELB), p50 derived from p105 and p52 derived from p100 [12]. The predominant species of this family are p65/p50 heterodimers, although other forms of homodimers and heterodimers also exist. The NF-κB/REL dimers are normally bound to inhibitor of kappa-B (IκB) isoforms under basal conditions. Phosphorylation of these IκB forms by IκB kinase leads to their ubiquitination and proteosomal degradation. Gene expression patterns induced by cytokines through NF-κB are reported to be predominantly pro-apoptotic in rat beta cells [13, 14]. Previous studies in other cell types have also reported cytokine-mediated induction of a number of anti-apoptotic genes including BIRC3, TRAF1 and TNFAIP3 through activation of NF-κB [15–17].
The objectives of the present study were to: (1) examine the global gene expression response of human islet preparations to a combination of IL1β, TNF-α and IFN-γ using microarrays; (2) elucidate a common biological pattern by combining single-gene analysis with exploration of pathways by means of gene set enrichment analysis (GSEA); and (3) determine the role of NF-κB in the synergistic induction of the promoter of BIRC3, a caspase inhibitor, by cytokines in MIN6 cells, a mouse beta cell line. These studies demonstrated the importance of NF-κB pathway in the induction of anti-apoptotic genes by proinflammatory cytokines in human islets and MIN6 cells. We postulate that cytokines not only exert deleterious effects on human islets but also act concurrently to maintain functional integrity by inducing genes related to cell survival.
Methods
Islet procurement and culture
Human islets were prepared by collagenase digestion by the Islet Cell Resource (ICR) Center at the University of Colorado at Denver using the Edmonton protocol (cold ischaemia time 4 to 9 h) (Approved by Institutional Review Board). All donors were brain-dead, heart-beating individuals from the state of Colorado who died in motor vehicle accidents. None had previous history of diabetes or inflammatory diseases. Islet purity (75–80%) and viability (76–96%) was determined by dithizone and Syto13/ethidium bromide staining respectively using standard operation procedures defined by the Clinical Islet Laboratory, SMRI, Edmonton, AB, Canada. Islets were precultured for 12 to 24 h in Miami media (CMRL 1066 supplemented media 99-603-CV; Mediatech, Hendon, VA, USA) containing 0.5% (wt/vol.) human serum albumin. Islets were exposed for 24 h to a mixture of IL1β (2 ng/ml; 100 U/ml), IFN-γ (10 ng/ml; 200 U/ml) and TNF-α (10 ng/ml; 1,000 U/ml) or IL1β (10 ng/ml; 500 U/ml), IFNγ (25 ng/ml; 500 U/ml) and TNFα (25 ng/ml; 2,500 U/ml) individually (Roche Applied Science, Indianapolis IN, USA) [18–20].
RNA isolation and microarray
Total RNA extraction, purification and labelling were performed as described previously [19]. Using standard Affymetrix protocol (Affymetrix, Santa Clara, CA, USA), 15 µg of biotin-labelled cRNA was hybridised to Human genome HG U133 Plus 2.0 microarray chips (Affymetrix) containing 54,675 probe sets representing around 22,000 unique genes.
Genome-wide expression and statistical analysis
The initial data analysis, quality control and normalisation were performed by GC Random-Multiple array analysis using Bioconductor Project software (www.bioconductor.org, accessed 3 March 2009) [21]. Probes were analysed with an alternative annotation package that (1) removes bad-quality or redundant probes [22], (2) discards about 30% of probes that do not reliably detect the expression of genes or align to more than one gene and (3) reduces the number of genes represented on Affymetrix HGU133 plus 2.0 chips to 17814. A permissive filtering was applied to each gene to include those genes that had an expression intensity of log2 (10) or higher in at least two conditions. Differential expression analysis was assessed by linear models and empirical Bayes moderated F statistics [23]. Genes were considered significant if adjusted p values (corrected by Benjamini and Hochberg’s procedure for multiple hypothesis testing) were below 0.1 [23].
Gene set enrichment analysis
Genome-wide expression profiles were divided into two classes (untreated and cytokine-treated) and compared with sets of genes that are grouped together in the same metabolic pathway or share similar Gene Ontology function derived from ten publicly available and manually curated databases (GSEA version 2.0, C2, Molecular Signature Database, MsigDB). The detailed mathematical description of the GSEA methodology [24, 25] and software can be found at www.broad.mit.edu/GSEA/ (accessed 3 March 2009).
BIRC3 promoter analysis by transient transfection
The promoter region of BIRC3 contains three NF-κB sites of which two (A and B) have been shown to be responsive to cytokines [26]. The following promoter constructs of BIRC3 linked to firefly luciferase reporter were generated as described earlier [15]: (1) the full-length promoter of BIRC3 (−1931 to +27); (2) truncated promoter with NF-κB sites (A and B) (−242 to +27); (3) truncated promoter with one NF-κB site (B); and (4) truncated promoter without NF-κB sites (−107 to +27). Plasmids expressing p65 and super-repressor of IκB (SR-IκB) were provided by T. Okamoto (Department of Molecular and Cellular Biology, Nagoya University, Nagoya, Japan) [27] and A. Rabson (Center for Advanced Biotechnology and Medicine, Piscataway, NJ, USA) [28] respectively. Transient transfections in MIN6 cells, a mouse pancreatic beta cell line (passage numbers 25–35), were carried out using a reagent (LipofectAMINE 2000; Invitrogen-Life Technologies, Carlsbad, CA, USA) [29]. A constitutively active renilla luciferase (pRL-TK-luc) was included to correct for transfection efficiency. After 6 h, the transfected cells were exposed to 1 ng/ml of IL1β (5 U/ng), 5 ng/ml of TNF-α (100 U/ng) and 5 ng/ml of IFN-γ 50 U/ng, alone or in combinations, for 24 h. These cytokine concentrations are comparable to those used in previous reports [30, 31]. Luciferase activity was measured in the cell lysates using a dual luciferase assay kit (Promega, Madison, WI, USA).
Real-time quantitative RT-PCR
Total RNA was isolated from cytokine-treated MIN6 cells using a kit (Versagene RNA isolation kit; Qiagen, Valencia, CA, USA). The mRNA levels of Birc3, Bcl2A1, Cflar, Tnfaip3, Traf1 and Fas were measured by real-time quantitative RT-PCR using Taqman probes as described [32]. The sequences of forward and reverse primers and fluorescently labelled probes are listed in Electronic supplementary material (ESM) Table 1. The mRNA levels for all genes were normalised to 18S ribosomal RNA. The expression of corresponding genes in human islets exposed to cytokines was determined by Assay on Demand (Applied Biosystems, Foster City, CA, USA) and normalised to HPRT1.
Immunocytochemistry
MIN6 cells were cultured on cover slips, fixed in 4% (wt/vol.) paraformaldehyde and washed with PBS. They were permeabilised for 90 min at room temperature with PBS containing 0.2% (vol./vol.) Triton X-100 and 5% (wt/vol.) BSA, followed by exposure to the primary antibody (anti-p65; 1:250) at 4°C overnight. The cells were washed in PBS, incubated in the presence of the secondary antibody linked to Cy3 (anti-rabbit) and DAPI (2 μg/ml; nuclear staining) for 90 min at room temperature. The cells were then washed in PBS, mounted on slides with mounting medium and examined by fluorescent microscopy.
Statistics
Statistical analysis was performed by one-way ANOVA with Dunnett’s multiple comparison test.
Results
Global gene expression patterns in cytokine-treated human islets
Affymetrix HG U133 Plus 2.0 gene chips that enable concurrent analysis of 54,675 probe sets were used to analyse global gene expression profile of human islet preparations exposed for 24 h to IL1β, TNF-α and IFN-γ. The islets exhibited profound changes in the expression of a number of genes, many of which have been previously documented either in cytokine-exposed rodent islets or pancreatic beta cells purified by flow cytometry [14, 33]. Taking into account alternative annotations, we obtained information on expression of 17,814 genes [22]. At a false discovery rate cut-off value of 0.1, 572 gene transcripts were upregulated (ESM Table 2) and 406 genes were downregulated by the cytokines (ESM Table 3). In addition to the induction of apoptotic genes including CASP7, BID, TNFRSF1B, FAS and TNF, a number of anti-apoptotic genes including BIRC3, BCL2A1, CFLAR, TNFAIP3 and TRAF1 were also upregulated.
Gene set enrichment analysis
To understand the biological pathways modulated by the cytokines, we focused on metabolic and signalling pathways compiled in GSEA and capable of revealing significant changes even though the average change per gene might only be 20% [25]. Some eight or more different pathways and gene sets indicated the enrichment of NF-κB and RELA transcripts, including TNF, MAPK8, NF-κB1A, TRAF2, IL6, CHUK, JAK2, STAT5A, RIPK1 and TRAF6. In addition, several gene pathways upregulated by type 1 (α, β) and type 2 (γ) interferon were also enriched, with IRF1, IFITM1, GIP2, TRIM21, MX1, MX2 OAS1 and TAP1 showing enrichment in several allied gene sets. Inflammation and propagation of inflammatory signals like the dendritic cell pathway (CSF2, TLR2, IL12A), cytokine pathway (TNF, IL1A, IL15, IL6, IL12A), inflammatory pathway (TNF, CSF1, CSF2, CSF3, HLA-DRA, IL15, IL1A, IL6, IL11, IL12A) and IL1 receptor pathway (TNF, IL1A, IL1B, RELA, IRAK2, IL1RN, IL6, MAPK8, NFKB1, MAP2K3, CHUK, NFKBIA, TRAF6, MYD88) were enriched. Several stress-related pathways were significantly enriched, including matrix metalloproteinases induction (TNF, MMP25, MMP10, MMP3, MMP12, MMP14, MMP2, TCF20, MMP9, MMP1) and inducible nitric oxide induction (NOS2A [also known as NOS2], JAK2, STAT4, IL12A, TYK2, CD3D) (GSEA supplementary data; available from www.uchsc.edu/misc/diabetes/Sarkar/ExtractedFiles/index.html, accessed 27 January 2009).
Validation of microarray data by quantitative PCR
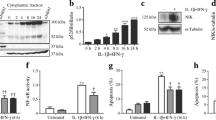
The upregulation of BIRC3, BCL2A1, CFLAR, TNFAIP3, TRAF1 and FAS by cytokines in human islets was validated by real-time RT-PCR (Fig. 1). A combination of cytokines (IL1β, TNF-α and IFN-γ) increased the expression of BCL2A1 by 11-fold and other genes by three to fourfold. When islets were exposed to individual cytokines, even at higher concentrations, upregulation of BIRC3, BCL2A1 and FAS by IL1β or TNF-α was moderate. However, no significant increase in the levels of CFLAR, TNFAIP3 and TRAF1 were observed when islets were exposed to individual cytokines. These findings suggest that the cytokines act synergistically to induce many of these genes. To determine whether cytokines induce these genes through NF-κB, we used Bay 11-7085, an inhibitor of IκB phosphorylation (Fig. 1g, h). NF-κB normally resides in the cytoplasm bound to the inhibitor, IκB. When IκB is phosphorylated and undergoes proteosomal degradation, NF-κB translocates to nucleus and drives the expression of its target genes. The addition of 7.5 μmol/l Bay 11-7085 to cultured islets decreased cytokine-mediated induction of BIRC3, BCL2A1, CFLAR, TRAF1, TNFAIP3 and FAS by 45% to 65% (p < 0.01) suggesting a role for NF-κB. Immunohistochemical localisation of baculovirus IAP repeat (BIR)3 was also noted in insulin-positive islet cells with cytokine exposure (ESM Fig. 1). Expression of these genes was also analysed in islets after 72 h of cytokine exposure. The upregulation of many of the anti-apoptotic genes by cytokines was completely lost at this time point (ESM Fig. 2), accompanied by increased cell death as noted by TUNEL staining (data not shown).
Gene expression analysis in cytokine-treated human islets. Human islets (3,000 islet equivalents [IEQ]) were cultured for 24 h in the presence of the indicated cytokines alone or in combination (Mix), and in the absence or presence of 7.5 μmol/l Bay 11-7085 (Bay). Total RNA was isolated and the mRNA levels of a BIRC3, b BCL2A1, c CFLAR, d TNFAIP3, e TRAF1 and f FAS were determined by quantitative RT-PCR using Taqman probes and normalised to HPRT1. The results are mean±SE. **p < 0.01 compared with untreated controls (C); † p < 0.01 vs a combination of cytokines. g, h Human islets (3,000 IEQ) were cultured in the presence of a combination of cytokines (Cyt), in the absence and presence of Bay 11-7085 (Bay) for 3 h. The treated islets were processed for immunoblot analysis g of phosphorylated IκBα. Blots were reprobed for beta actin. The bands were quantitated h by scanning densitometry and corrected for beta actin. ***p < 0.001 compared with untreated controls; ‡ p < 0.001 vs a combination of cytokines. C, control; IF, IFN-γ; IL, IL1-β; TN, TNF-α
NF-κB-dependent induction of anti-apoptotic genes in MIN6 cells
To determine whether cytokine-mediated induction of anti-apoptotic genes was not restricted to human islets alone, we measured the mRNA levels of these genes in MIN6 cells exposed to a mixture of cytokines for 6 to 24 h (Fig. 2). A time-course of induction of these genes by cytokines showed ten- to 23-fold induction of Birc3, Tnfaip3 and Fas by 6 h and a gradual decrease in induction by 12 and 24 h. The induction of Cflar and Traf1 on the other hand was sustained for 24 h. Cytokine-mediated induction of these genes was decreased by 40% to 50% after pre-incubation with Bay 11-7085. Unlike in human islets, cytokines did not significantly induce Bcl2a1 in MIN6 cells.
Gene expression analysis in cytokine-treated MIN6 cells. MIN6 cells cultured in 100 mm dishes were exposed to a combination of IL1β (1 ng/ml), TNF-α (5 ng/ml) and IFN-γ (5 ng/ml) in the absence or presence of 10 μmol/l Bay 11-7085. Total RNA was isolated from cytokine-treated MIN6 cells and the mRNA levels of a Birc3, b Bcl2a1, c Cflar, d Tnfaip3, e Traf1 and f Fas were measured by real-time quantitative RT-PCR and normalised to 18S ribosomal RNA. The results are mean±SE of four independent experiments. ***p < 0.001 compared with untreated controls; † p < 0.001 vs cytokines. White bars, control; grey bars, Bay 11-7085; black bars, cytokines; hatched bars, cytokines and Bay 11-7085
Induction of BIRC3 promoter by TNF-α and IL1β
BIRC3, a caspase inhibitor, belongs to the family of inhibitors of apoptosis (IAP), which play a role in cellular recovery from apoptosis [34]. Therefore, to determine whether NF-κB facilitates this process, the regulation of BIRC3 expression by cytokines was further examined at the promoter level by transient transfection in MIN6 cells. The effects of individual cytokines on the activity of human BIRC3 promoter linked to a luciferase reporter construct are depicted in ESM Fig. 3. TNF-α was found to induce the reporter in a dose-dependent manner, up to fourfold at 20 ng/ml. IL1β was a weaker inducer with a twofold increase at 1 ng/ml. IFN-γ did not induce BIRC3 promoter activity significantly even at 20 ng/ml. The induction of BIRC3 by cytokines may be cell type-dependant, as a previous study reported stronger induction by cytokines, especially by IL1β in HeLa cells [26] and by TNF-α in H441 and A549 pulmonary epithelial cells, but not in U937 cells [35].
Synergy between cytokines in the induction of BIRC3 promoter
Induction of BIRC3 promoter by TNF-α was further potentiated by IL1β (55%; p < 0.001). IFN-γ did not induce BIRC3 promoter on its own; however, it enhanced TNF-α action significantly (p < 0.001) by 158%. A combination of IL1β, TNF-α and IFN-γ showed around 700% induction, a value that is significantly (p < 0.001) greater than the sum of effects seen with individual cytokines, suggesting a synergy between them. Cytokine-mediated induction of a truncated BIRC3 promoter that retained both NF-κB response elements (A and B) was similar to that of the full-length promoter (Fig. 3a,b). Deletion of one of the response elements (A) resulted in partial loss of induction (Fig. 3c). Cytokines did not induce BIRC3 promoter significantly when both NF-κB response elements (A and B) were deleted (Fig. 3d).
Induction of BIRC3 promoter by cytokines through NF-κB responsive sites. MIN6 cells cultured to ~70% confluence were transfected with a luciferase reporter gene driven by a the full-length BIRC3 promoter (−1931 to +27) or truncated promoters with b two (A and B) NF-κB sites (−242 to +27), c one (B) NF-κB site (−205 to +27) or d no NF-κB site (−107 to +27) of BIRC3 gene, along with constitutively active renilla luciferase (for transfection efficiency). After 6 h of transfection, the cells were exposed to cytokines IL1β (IL; 1 ng/ml), TNF-α (TN; 5 ng/ml) or IFN-γ (IF; 5 ng/ml) alone or in different combinations. Firefly and renilla luciferase activities were determined 18 h later using a dual luciferase assay kit (Promega). The ratio of the two luciferase activities was taken as the activity of the promoter construct being examined. The results are mean±SE of six independent experiments. ***p < 0.001 vs control; † p < 0.001 compared with indicated individual cytokines. C, control; IF, IFN-γ; IL, IL1-β; TN, TNF-α
NF-κB-mediated induction of BIRC3 promoter by cytokines
To confirm the role of NF-κB in cytokine action, two methods were used to inhibit this pathway. First, the addition of Bay 11-7085, an inhibitor of IκB phosphorylation, decreased induction of BIRC3 by cytokines (Fig. 4a). Second, cotransfection with SR-IκB, a mutant of IκB that cannot be phosphorylated, resulted in significant blocking of induction by cytokines (Fig. 4b). Overproduction of the p65 subunit of NF-κB complex resulted in increased basal promoter activity of BIRC3 by 2.5-fold, but subsequent exposure to cytokines did not lead to additional induction. We also observed that the nuclear localisation of p65 was inhibited by Bay 11-7085 (Fig. 4c).
Induction of BIRC3 promoter by cytokines through NF-κB. a MIN6 cells transfected with BIRC3-luc were exposed for 24 h to a mixture of cytokines (IL1β 1 ng/ml, TNF-α 5 ng/ml, IFN-γ 5 ng/ml) in the absence or presence of increasing concentrations of Bay 11-7085 (Bay). Firefly and renilla luciferase activities were determined in the cell lysates. The values represent mean±SE of four independent experiments. ***p < 0.001 vs cytokines without Bay. b MIN6 cells were transfected with BIRC3-luc and p65 or SR-IκB or vector. Transfected cells were exposed to a combination (Cyt) of IL 1-β (1 ng/ml), TNF-α (5 ng/ml) and IFN-γ (5 ng/ml) for 24 h and luciferase activities were determined. ***p < 0.001 compared with untreated controls; † p < 0.001 vs untreated vector control. c MIN6 cells cultured on coverslips were exposed to Bay 11-7085 (Bay) or a mixture of cytokines or both for 3 h. Treated cells were fixed in 4% paraformaldehyde, permeabilised and immunostained for p65 (Cy3; red). The nuclei were stained with DAPI (blue). Images were examined by fluorescent microscopy
Activation of caspase-3 by cytokines
To determine the effects of cytokine treatment on the survival of MIN6 cells, activation of caspase-3, a marker of apoptosis, was examined. When the MIN6 cells were exposed to individual cytokines for 24 h, TNF-α alone activated caspase-3, its action being potentiated by IL1β and IFN-γ (Fig. 5a). A combination of all three cytokines caused maximum activation of caspase-3. When MIN6 cells were incubated in the presence of a combination of cytokines for 6 to 24 h, significant activation of caspase-3 was seen by 18 h, which increased further at 24 h (Fig. 5b). However, no overt apoptotic cell death was observed by TUNEL assay (results not shown).
Activation of caspase-3 by cytokines. a MIN6 cells were exposed for 24 h to cytokines IL1β (IL 1 ng/ml), TNF-α (TN 5 ng/ml) or IFN-γ (IF 5 ng/ml) alone or in different combinations. b MIN6 cells were incubated in the absence or presence of a combination of cytokines for 6 to 24 h. Black bars, control; hatched bars, cytokines. The treated cells (a, b) were processed for immunoblot analysis of active cleaved form of caspase-3 and blots reprobed for beta actin. Bands were quantitated by scanning densitometry and corrected for beta actin. Representative blots are shown. **p < 0.01, ***p < 0.001 vs untreated control. C, control; IF, IFN-γ; IL, IL1-β; TN, TNF-α
Discussion
The proinflammatory cytokines IL1β, TNF-α and IFN-γ play an important role in beta cell apoptosis in autoimmune diabetes. A large body of in vitro experiments suggests that cytokine-induced NF-κB activation is an important signalling event in triggering beta cell apoptosis [13, 14, 36, 37]. NF-κB has been suggested to be pro-apoptotic in beta cells, whereas it is anti-apoptotic in other cell types [16, 38]. By combining global gene analysis via GSEA and quantitative RT-PCR analysis, we demonstrate here that cytokines upregulate several anti-apoptotic genes, including BIRC3, BCL2A1, CFLAR, TNFIAP3 and TRAF1, through NF-κB-mediated signalling in human islets. We also demonstrate that the cytokines induce the promoter of anti-apoptotic Birc3 through NF-κB activation synergistically in MIN6 cells, a mouse beta cell line.
Increased expression of NF-κB1, NF-κB2 and RELA subunits of REL/NF-κB transcription factors in cytokine-treated human islets was observed. The target genes of NF-κB as follows were upregulated by exposure of human islets to a combination of IL1β, TNF-α and IFN-γ: (1) cytokines/chemokines and their modulators (IL1α, IL1β, IL6, IL8, IP10, CXCL1, lymphotoxin β, MIP1α, MIP-2, RANTES, TNFα); (2) immunoreceptors (β2 microglobulin); (3) antigen presentation (TAP1); (4) cell adhesion (ICAM1); (5) acute-phase proteins (complement factor B, urokinase-type plasminogen activator, COX2); (6) cell surface receptors (LOX1); and (7) growth factors and their modulators (G-CSF, GM-CSF, CSF-1). This pattern of upregulation was largely consistent with inflammation. Gene expression profiles of human islets treated with IL1β plus IFN-γ [39] and INF-γ alone [19] have been previously reported. This is the first report to show that exposure of cultured human islets to a combination of IL1β, TNF-α and IFNγ results in upregulation of several anti-apoptotic genes via NF-κB, namely BIRC3, BCL2A1, CFLAR, TNFAIP3 and TRAF1 (Table 1). Expression of these genes was confirmed by real-time quantitative RT-PCR using Taqman probes (Fig. 1). Among these genes, cytokine-mediated induction of TNFAIP3 alone has been reported previously in human islets [40]. The probable reason for our findings on other anti-apoptotic genes could be the use of a combination of IL1β, TNF-α and IFN-γ. IL1β and TNF-α are known to activate NF-κB whereas IFN-γ acts primarily through Janus kinase-mediated activation of the transcription factor STAT-1 [41]. In the present study, significant induction of many of these genes was seen only when a combination of three cytokines were used (Fig. 1). We also observed that IFN-γ enhanced induction of BIRC3 promoter by TNF-α through activation of NF-κB (Fig. 4). The effects of IFN-γ were lost after deletion of NF-κB response elements in BIRC3 promoter. Additionally, maximum induction of BIRC3 promoter was observed when all three cytokines were combined, whereas this synergistic effect was lost when MIN6 cells were cotransfected with IκBα super repressor and by the inhibitor Bay 11-7085.
The molecular mechanism of beta cell death by apoptosis is not fully understood. The products of anti-apoptotic genes induced by NF-κB signalling (Fig. 1) are known to promote cell survival by acting at several critical steps in the extrinsic and intrinsic pathways of apoptosis. In the extrinsic pathway, the death receptors, when bound to ligands, recruit the adaptor protein Fas-associated death domain (FADD), which in turn recruits caspase-8 to form the death-inducing signalling complex (DISC). Caspase-8 and FADD-like apoptosis regulator (CFLAR) binds to FADD within DISC and inhibits caspase-8 activation. TNF receptor-mediated signalling, which also leads to caspase-8, is inhibited by TNF-α-induced protein 3 (TNFAIP3) and TNF receptor-associated factor 1 (TRAF1), a member of TRAFs family. TNFAIP3 has been shown to protect a mouse beta cell line and rat islets from cytokines [17, 40]. The intrinsic pathway of apoptosis is regulated by the pro- and anti-apoptotic B-cell lymphoma 2 (BCL2) family of proteins. The anti-apoptotic BCL2-related protein A1 (BCL2A1) inhibits the release of cytochrome C, which activates caspase-9. BIRC3, a caspase inhibitor, inhibits both pathways of apoptosis. The IAP are a conserved family of proteins that inhibit caspases and play a role in cellular recovery from apoptosis [34]. The IAP family is characterised by the presence of one or more 70 to 80 AA BIR domains and in humans include BIRC1, BIRC2, BIRC3, BIRC4, BIRC5, BIRC6, BIRC7 and BIRC8. Equilibrium between apoptosis-inducing caspases and IAPs is an important checkpoint during the induction of apoptosis. BIRC3 triggers the proteosomal degradation of caspases by binding to them through the really interesting new gene domain [34]. Thus NF-κB-regulated genes provide multiple checkpoints when apoptosis is induced as a cytoprotective response.
A considerable amount of microarray data on cytokine-mediated NF-κB-regulated gene expression patterns in beta cells are available from the reports of Eizirik and co-workers [14, 37, 42]. These studies suggest that NF-κB-mediated induction of pro-apoptotic genes predominantly mediates beta cell death in type 1 diabetes. Some of these pro-apoptotic genes could exert delayed indirect effects. For example, NF-κB-dependent inducible nitric oxide synthase generates nitric oxide, which causes beta cell dysfunction and death [43]. Furthermore, FAS-mediated extrinsic pathway of apoptosis interacts with the intrinsic mitochondrial pathway through generation of truncated Bid, which induces the release of cytochrome C from the mitochondria [44]. Cytokines could also induce apoptosis through NF-κB-independent pathways including activation of c jun N-terminal kinase [45]. Thus, beta cell death induced by cytokines could result from late events triggered by NF-κB-regulated pro-apoptotic genes and NF-κB-independent signalling pathways. Considering the complex nature of cytokine-mediated signalling, it is difficult, on the basis of the array of genes induced, to ascertain that NF-κB is predominantly pro-apoptotic. Although NF-κB-mediated protective pathways may seem to be transient or overshadowed by the pro-apoptotic response to cytokines, they can be critically exploited both in vivo and in vitro to sustain islet survival.
Widespread overt apoptosis measured by TUNEL assay (data not shown) was not evident in cytokine-treated human islets or MIN6 cells at 24 h. We did, however, observe the activation of caspase-3 by 18 to 24 h in MIN6 cells (Fig. 5). It has been suggested that intervention downstream of caspase activation could allow functional recovery through IAP families [34]. A time-course of gene expression analysis (Fig. 2) in MIN6 cells exposed to cytokines revealed a stronger induction of anti-apoptotic genes in the early phase up to 6 h, which is not sustained over time. For example, Birc3 and Tnfaip3 were induced by 10- to 20-fold at 6 h compared with three- to eightfold induction at 24 h, suggesting that initially pro- and anti-apoptotic pathways are both induced. However, after continued exposure to cytokines, the cell death pathway seems to prevail. As such, autoimmune destruction of beta cells can be seen as a slow process, with induction of anti-apoptotic genes possibly playing a role in prolonging beta cell survival by opposing the effects of the pro-apoptotic pathway.
Previous studies have reported conflicting results on beta cell survival after blocking NF-κB activation. For example, adenoviral transduction of rat beta cells [36] and human islets [46] with NF-κB repressor leads to inhibition of cytokine-induced apoptosis suggesting a pro-apoptotic role for this transcription factor. A recent study demonstrated in an inducible transgenic mouse model that beta cell-specific inhibition of NF-κB results in protection against low-dose streptozotocin-induced diabetes [47]. In contrast, accelerated development of autoimmune diabetes has been reported in transgenic NOD mice expressing a repressor of NF-κB in beta cells [48]. Another study observed that inhibition of NF-κB sensitises cultured beta cells to TNF-α-mediated apoptosis [49]. These reports and our current findings suggest that by selective inhibition of the pro-apoptotic effects of NF-κB, therapeutic strategy in type 1 diabetes could be further improved. Future studies should examine the effects of the members of NF-κB/IκB families on gene targets to determine if such selective modulation is feasible.
Abbreviations
- BCL2:
-
B-cell lymphoma 2
- BCL2A1:
-
BCL2-related protein A1
- BIR:
-
Baculovirus IAP repeat
- CFLAR:
-
Caspase-8 and FADD-like apoptosis regulator
- DISC:
-
Death-inducing signalling complex
- FADD:
-
Fas-associated death domain
- GSEA:
-
Gene set enrichment analysis
- IAP:
-
Inhibitors of apoptosis
- IκB:
-
Inhibitor of kappa-B
- NF-κB:
-
Nuclear factor kappa-B
- SR-IκB:
-
Super-repressor of IκB
- TNFAIP3:
-
TNF-α-induced protein 3
- TRAF1:
-
TNF receptor-associated factor 1
References
Rosmalen JG, Leenen PJ, Pelegri C, Drexhage HA, Homo-Delarche F (2002) Islet abnormalities in the pathogenesis of autoimmune diabetes. Trends Endocrinol Metab 13:209–214
Yoon JW, Jun HS (2001) Cellular and molecular pathogenic mechanisms of insulin-dependent diabetes mellitus. Ann N Y Acad Sci 928:200–211
Delovitch TL, Singh B (1997) The nonobese diabetic mouse as a model of autoimmune diabetes: immune dysregulation gets the NOD. Immunity 7:727–738
Mandrup-Poulsen T (1996) The role of interleukin-1 in the pathogenesis of IDDM. Diabetologia 39:1005–1029
Delaney CA, Pavlovic D, Hoorens A, Pipeleers DG, Eizirik DL (1997) Cytokines induce deoxyribonucleic acid strand breaks and apoptosis in human pancreatic islet cells. Endocrinology 138:2610–2614
Andre I, Gonzalez A, Wang B, Katz J, Benoist C, Mathis D (1996) Checkpoints in the progression of autoimmune disease: lessons from diabetes models. Proc Natl Acad Sci U S A 93:2260–2263
Moriwaki M, Itoh N, Miyagawa J et al (1999) Fas and Fas ligand expression in inflamed islets in pancreas sections of patients with recent-onset Type I diabetes mellitus. Diabetologia 42:1332–1340
Mauricio D, Mandrup-Poulsen T (1998) Apoptosis and the pathogenesis of IDDM: a question of life and death. Diabetes Metab Rev 47:1537–1543
Pavlovic D, Chen MC, Gysemans CA, Mathieu C, Eizirik DL (1999) The role of interferon regulatory factor-1 in cytokine-induced mRNA expression and cell death in murine pancreatic beta-cells. Eur Cytokine Netw 10:403–412
Hoorens A, Stange G, Pavlovic D, Pipeleers D (2001) Distinction between interleukin-1-induced necrosis and apoptosis of islet cells. Diabetes 50:551–557
Luo JL, Kamata H, Karin M (2005) IKK/NF-kappaB signaling: balancing life and death—a new approach to cancer therapy. J Clin Invest 115:2625–2632
Pahl HL (1999) Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18:6853–6866
Ortis F, Pirot P, Naamane N et al (2008) Induction of nuclear factor-kappaB and its downstream genes by TNF-alpha and IL-1beta has a pro-apoptotic role in pancreatic beta cells. Diabetologia 51:1213–1225
Cardozo AK, Heimberg H, Heremans Y et al (2001) A comprehensive analysis of cytokine-induced and nuclear factor-kappa B-dependent genes in primary rat pancreatic beta-cells. J Biol Chem 276:48879–48886
Ueda T, Akiyama N, Sai H et al (2001) c-IAP2 is induced by ionizing radiation through NF-kappaB binding sites. FEBS Lett 491:40–44
Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS Jr (1998) NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 281:1680–1683
Liuwantara D, Elliot M, Smith MW et al (2006) Nuclear factor-kappaB regulates beta-cell death: a critical role for A20 in beta-cell protection. Diabetes 55:2491–2501
Oetjen E, Blume R, Cierny I et al (2007) Inhibition of MafA transcriptional activity and human insulin gene transcription by interleukin-1beta and mitogen-activated protein kinase kinase kinase in pancreatic islet beta cells. Diabetologia 50:1678–1687
Sarkar SA, Wong R, Hackl SI et al (2007) Induction of indoleamine 2,3-dioxygenase by interferon-gamma in human islets. Diabetes 56:72–79
Mokhtari D, Myers JW, Welsh N (2008) MAPK kinase kinase-1 is essential for cytokine-induced c-Jun NH2-terminal kinase and nuclear factor-kappaB activation in human pancreatic islet cells. Diabetes 57:1896–1904
Gentleman RC, Carey VJ, Bates DM et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80
Dai M, Wang P, Boyd AD et al (2005) Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 33:e175
Smyth GK, Michaud J, Scott HS (2005) Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21:2067–2075
Subramanian A, Tamayo P, Mootha VK et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550
Mootha VK, Lindgren CM, Eriksson KF et al (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273
Hong SY, Yoon WH, Park JH, Kang SG, Ahn JH, Lee TH (2000) Involvement of two NF-kappa B binding elements in tumor necrosis factor alpha -, CD40-, and Epstein–Barr virus latent membrane protein 1-mediated induction of the cellular inhibitor of apoptosis protein 2 gene. J Biol Chem 275:18022–18028
Jiang X, Takahashi N, Matsui N, Tetsuka T, Okamoto T (2003) The NF-kappa B activation in lymphotoxin beta receptor signaling depends on the phosphorylation of p65 at serine 536. J Biol Chem 278:919–926
Suh J, Payvandi F, Edelstein LC et al (2002) Mechanisms of constitutive NF-kappaB activation in human prostate cancer cells. Prostate 52:183–200
Jambal P, Masterson S, Nesterova A et al (2003) Cytokine-mediated downregulation of the transcription factor CREB in pancreatic beta -cells. J Biol Chem 278:23055–23065
Cardozo AK, Proost P, Gysemans C, Chen MC, Mathieu C, Eizirik DL (2003) IL-1beta and IFN-gamma induce the expression of diverse chemokines and IL-15 in human and rat pancreatic islet cells, and in islets from pre-diabetic NOD mice. Diabetologia 46:255–266
Bonny C, Oberson A, Steinmann M, Schorderet DF, Nicod P, Waeber G (2000) IB1 reduces cytokine-induced apoptosis of insulin-secreting cells. J Biol Chem 275:16466–16472
Pugazhenthi S, Nesterova A, Sable C et al (2000) Akt/Protein kinase B up-regulates Bcl-2 expression through cAMP-response element-binding protein. J Biol Chem 275:10761–10766
Cardozo AK, Kruhoffer M, Leeman R, Orntoft T, Eizirik DL (2001) Identification of novel cytokine-induced genes in pancreatic beta-cells by high-density oligonucleotide arrays. Diabetes 50:909–920
Liston P, Fong WG, Korneluk RG (2003) The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene 22:8568–8580
Pryhuber GS, Huyck HL, Staversky RJ, Finkelstein JN, O'Reilly MA (2000) Tumor necrosis factor-alpha-induced lung cell expression of antiapoptotic genes TRAF1 and cIAP2. Am J Respir Cell Mol Biol 22:150–156
Heimberg H, Heremans Y, Jobin C et al (2001) Inhibition of cytokine-induced NF-kappaB activation by adenovirus-mediated expression of a NF-kappaB super-repressor prevents beta-cell apoptosis. Diabetes 50:2219–2224
Kutlu B, Cardozo AK, Darville MI et al (2003) Discovery of gene networks regulating cytokine-induced dysfunction and apoptosis in insulin-producing INS-1 cells. Diabetes 52:2701–2719
Beg AA, Baltimore D (1996) An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science 274:782–784
Ylipaasto P, Kutlu B, Rasilainen S et al (2005) Global profiling of coxsackievirus- and cytokine-induced gene expression in human pancreatic islets. Diabetologia 48:1510–1522
Grey ST, Arvelo MB, Hasenkamp W, Bach FH, Ferran C (1999) A20 inhibits cytokine-induced apoptosis and nuclear factor kappaB-dependent gene activation in islets. J Exp Med 190:1135–1146
Ihle JN (1995) Cytokine receptor signalling. Nature 377:591–594
Eizirik DL, Kutlu B, Rasschaert J, Darville M, Cardozo AK (2003) Use of microarray analysis to unveil transcription factor and gene networks contributing to beta cell dysfunction and apoptosis. Ann N Y Acad Sci 1005:55–74
Mandrup-Poulsen T (2001) Beta-cell apoptosis: stimuli and signaling. Diabetes 50(Suppl.1):S58–S63
Luo X, Budihardjo I, Zou H, Slaughter C, Wang X (1998) Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94:481–490
Bonny C, Oberson A, Negri S, Sauser C, Schorderet DF (2001) Cell-permeable peptide inhibitors of JNK: novel blockers of beta-cell death. Diabetes 50:77–82
Giannoukakis N, Mi Z, Rudert WA, Gambotto A, Trucco M, Robbins P (2000) Prevention of beta cell dysfunction and apoptosis activation in human islets by adenoviral gene transfer of the insulin-like growth factor I. Gene Ther 7:2015–2022
Eldor R, Yeffet A, Baum K et al (2006) Conditional and specific NF-kappaB blockade protects pancreatic beta cells from diabetogenic agents. Proc Natl Acad Sci U S A 103:5072–5077
Kim S, Millet I, Kim HS et al (2007) NF-kappa B prevents beta cell death and autoimmune diabetes in NOD mice. Proc Natl Acad Sci U S A 104:1913–1918
Chang I, Kim S, Kim JY et al (2003) Nuclear factor kappaB protects pancreatic beta-cells from tumor necrosis factor-alpha-mediated apoptosis. Diabetes 52:1169–1175
Acknowledgements
These studies were supported by Juvenile Diabetes Research Foundation grants (5-2005-1104 to S. Pugazhenthi, 02-05-60294 to J. C. Hutton and 1-2008-1021 to S. A. Sarkar), American Diabetes Association grant (1-06-JF-40 to S. Pugazhenthi). S. A. Sarkar is also supported by NIH/NIDDK K01 DK080193 and a NIDDK/DERC Pilot and Feasibility Grant. Pancreatic islets and core resources were provided by the University of Colorado Denver (NIH 5 U42 RR016599), NIH/NIDDK/JDRF Islet Cell Resource (ICR) Human Islet Distribution Program and the DERC Molecular and Bioinformatics core facilities (NIH P30 DK57516) to J. C. Hutton. The authors would like to thank J. A. Walters and C. Patel (Barbara Davis Center, University of Colorado Denver) for their assistance in the preparation of this manuscript. We thank R. Bouchard at the Denver VAMC-Digital Deconvolution Microscopy Core facility and U. Pugazhenthi at the University of Colorado Cancer Center-RT-PCR Core Facility for providing excellent technical support.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM Fig. 1
Induction of BIRC3 production in beta cells of cytokine-treated human islets. Islets were incubated in the presence of a mixture of cytokines (IL1β, TNF-α and IFN-γ) for 24 h and embedded in optimal cutting temperature compound. Frozen sections (7 μm thick) were incubated with guinea pig anti-insulin antibody and rabbit BIRC3 antibody and stained with secondary antibodies linked to FITC (insulin) and Cy3 (BIRC3) (PDF 3.21 kb)
ESM Fig. 2
Decreased expression of anti-apoptotic genes in human islets after 72 h exposure to cytokines. Human islets (3,000 islet equivalents [IEQ]) were cultured in the presence of indicated cytokines alone or in combination (Mix) for 72 h. Total RNA was isolated and the mRNA levels of (a) BIRC3, (b) BCL2A1, (c) CFLAR, (d) TNFAIP3, (e) TRAF1 and (f) FAS were determined by quantitative RT-PCR using Taqman probes and normalised to HPRT1. The results are represented as mean±SE. # p < 0.05, *p < 0.01 compared with untreated controls (PDF 808 kb)
ESM Fig. 3
Induction of BIRC3 promoter by IL1β, TNF-α. MIN6 cells were transfected with a luciferase reporter gene driven by the full-length promoter construct of BIRC3 and constitutively active renilla luciferase to correct for transfection efficiency. After 6 h of transfection, the cells were exposed to the indicated concentrations of IL1β (a), TNF-α (b) or IFN-γ (c) and processed 18 h later for luciferase assays. The values represent mean±SE of four independent experiments. *p < 0.01, **p < 0.001 compared with untreated controls. C, control; IF, IFN-γ; IL, ILF-β; TN, TNF-α (PDF 524 kb)
ESM Table 1
Primers and probes for real-time RT-PCR of mouse genes (Fig. 2) (PDF 26.8 kb)
ESM Table 2
Gene transcripts that were upregulated in human islets exposed for 24 h to cytokine mixture (IL1β, TNFα, IFNγ) (PDF 428 kb)
ESM Table 3
Gene transcripts downregulated (410) in human islets following 24 h exposure to cytokine mixture (IL1β, TNFα, IFNγ) (PDF 312 kb)
Rights and permissions
About this article
Cite this article
Sarkar, S.A., Kutlu, B., Velmurugan, K. et al. Cytokine-mediated induction of anti-apoptotic genes that are linked to nuclear factor kappa-B (NF-κB) signalling in human islets and in a mouse beta cell line. Diabetologia 52, 1092–1101 (2009). https://doi.org/10.1007/s00125-009-1331-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-009-1331-x