Abstract
Key message
The meiotic recombination gene Dmc1 on wheat chromosome 5D has been identified as a candidate for the maintenance of normal chromosome synapsis and crossover at low and possibly high temperatures.
Abstract
We initially assessed the effects of low temperature on meiotic chromosome synapsis and crossover formation in the hexaploid wheat (Triticum aestivum L.) variety ‘Chinese Spring’. At low temperatures, asynapsis and chromosome univalence have been observed before in Chinese Spring lines lacking the long arm of chromosome 5D (5DL), which led to the proposal that 5DL carries a gene (Ltp1) that stabilises wheat chromosome pairing at low temperatures. In the current study, Chinese Spring wild type and 5DL interstitial deletion mutant plants were exposed to low temperature in a controlled environment room during a period from premeiotic interphase to early meiosis I. A 5DL deletion mutant was identified whose meiotic chromosomes exhibit extremely high levels of asynapsis and chromosome univalence at metaphase I after 7 days at 13 °C, suggesting that Ltp1 is deleted in this mutant. Immunolocalisation of the meiotic proteins ASY1 and ZYP1 on ltp1 mutants showed that low temperature results in a failure to complete synapsis at pachytene. KASP genotyping revealed that the ltp1 mutant has a 4-Mb deletion in 5DL. Of 41 genes within this deletion region, the strongest candidate for the stabilisation of chromosome pairing at low temperatures is the meiotic recombination gene Dmc1. The ltp1 mutants were subsequently treated at 30 °C for 24 h during meiosis and exhibited a reduced number of crossovers and increased univalence, though to a lesser extent than at 13 °C. We therefore renamed our ltp1 mutant ‘ttmei1’ (temperature-tolerant meiosis 1) to reflect this additional loss of high temperature tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In plants, male reproductive development is highly sensitive to adverse environmental conditions including high and low temperatures (De Storme and Geelen 2014). In wheat, the reproductive phase is more sensitive to high temperatures than the vegetative phase (Fischer and Maurer 1976). This is also the case for low temperatures, with meiosis I identified as the most sensitive stage (Thakur et al. 2010). High temperatures during reproductive development can have a negative impact on grain yield (Fischer and Maurer 1976; Fischer 1985; Wardlaw et al. 1989), and even short periods of a moderately high temperature (20–24 h at 30 °C) during meiosis can reduce grain number (Saini and Aspinall 1982; Draeger and Moore 2017). Grain yield is also reduced when low temperatures occur during the booting stage (Ji et al. 2017), which broadly corresponds to meiosis (Barber et al. 2015).
In cereal crops such as wheat, where grains are an important yield factor, stress-induced male sterility generally has a negative effect on crop yield and performance (De Storme and Geelen 2014). In the UK and France in the mid-1980s, cold and wet weather during floral development in the wheat variety ‘Moulin’ caused significant sterility, resulting in a reduction in grain yield of more than 70% (Law 1999). The dramatic falls in yield were thought to be a result of reduced fertility caused by the occurrence of low temperatures at meiosis. It was suggested that Moulin might carry specific alleles from diverse sources that made the wheat more sensitive to cold weather during meiosis.
Meiosis is essential for gamete formation in sexually reproducing organisms. In early meiosis, homologous chromosomes align next to each other as pairs, and then, during synapsis, the pairs of homologs become linked tightly with each other through the polymerisation of a protein structure called the synaptonemal complex (SC), which assembles between the paired chromosomes (Page and Hawley 2004). The SC has a ladder-like structure consisting of two chromosome axes and a central region. Its assembly can be tracked by immunolocalisation of the meiotic proteins ASY1, which interacts with the chromosome axes (Boden et al. 2009), and ZYP1, which is associated with the central region (Higgins et al. 2005; Khoo et al. 2012). During zygotene, the axes of the two homologs begin to become connected by the assembly of the central region between them. The axes are now called lateral elements. At pachytene, the central structure links the homologs along their entire lengths and synapsis is completed. SC assembly is a highly temperature-sensitive process (Bilgir et al. 2013).
The SC is thought to provide the structural framework for meiotic recombination to take place. During recombination, at least one crossover (CO) must form between each pair of homologs, to forge a physical connection between the chromosomes. These physical connections can be seen cytologically and are called chiasmata. COs enable genetic information to be exchanged between chromosomes and are also needed for accurate chromosome segregation and balanced gametes in the daughter cells. Once COs have fully formed (at pachytene), the SC is disassembled (at diplotene), at which point the homologs are only connected via their chiasmata. They remain connected until the chromosomes segregate at anaphase I.
In bread wheat, which is an allopolyploid with three homeologous (related) diploid genomes (AABBDD), COs only form between homologs and not between homeologs, ensuring that the three genomes behave as diploids during meiosis. Normally, at metaphase I, only ring bivalents (with two chiasmata) and occasional rod bivalents (one chiasma) are present. At metaphase I, the 21 bivalents align on the equatorial plate before segregating at anaphase I.
Correct pairing and segregation of homologs is vital for maintaining the stability and fertility of the genome. Errors in meiosis can lead to aneuploidy or infertility. High and low temperatures can induce a variety of meiotic aberrations in plants including changes in the frequency of chiasma formation (CO frequency) (Elliott 1955; Dowrick 1957; Bayliss and Riley 1972a; Higgins et al. 2012). Reduction in chiasma formation at extremes of temperature is linked to disruption of chromosome synapsis. This can result in unpaired univalent chromosomes that segregate randomly during meiosis I or are lost completely (Bomblies et al. 2015). Temperature-associated synapsis failure has been reported in plants at both low and high temperatures, but the temperature at which meiosis fails varies in different species (reviewed in Bomblies et al. 2015). These temperature thresholds can also vary within a species where there are genotypic differences (Riley et al. 1966).
Chinese Spring is one of the more heat-sensitive wheat cultivars (Qin et al. 2008) and has been widely used to study the frequency of chiasma formation at low and high temperatures because different Chinese Spring genotypes respond differently to temperature extremes. At low temperatures, a reduction in chiasma frequency and an increase in chromosome univalence have been observed in Chinese Spring nullisomic 5D-tetrasomic 5B (N5DT5B) plants, which lack chromosome 5D and have two extra copies of 5B (Riley 1966; Bayliss and Riley 1972a). Wheat has an optimum temperature range of around 17–23 °C over the course of a growing season (Porter and Gawith 1999). In N5DT5B plants, the frequency of chiasma formation is progressively reduced as the temperature rises above or falls below the optimum range (Bayliss and Riley 1972a). At 15 °C, chiasma frequency is greatly reduced in N5DT5B plants (Riley 1966), and they exhibit pronounced chromosome pairing failure at 12 °C, which leads to complete male sterility (Hayter and Riley 1967). The reduced chiasma frequencies seen in N5DT5B plants at low temperatures occur because the chromosomes fail to pair during zygotene, though the temperature-sensitive phase was found to be during premeiotic interphase, prior to DNA synthesis (Bayliss and Riley 1972b).
The asynapsis observed in N5DT5B is also seen in plants that are nullisomic for 5D (40 chromosomes), but in plants that are tetrasomic for 5B (44 chromosomes) synapsis is normal (Riley et al. 1966), so it was inferred that the asynapsis in N5DT5B could not be a result of the extra dosage of 5B, but that there must be a gene on chromosome 5D that stabilises chromosome pairing at low temperatures. This gene was named low-temperature pairing (Ltp) by Hayter and Riley (1967). Ltp was further defined to the long arm of chromosome 5D (Hayter 1969) and later renamed Ltp1 (Queiroz et al. 1991), but its exact location has never been mapped. It has been suggested that chiasma frequency also falls progressively in N5DT5B plants at temperatures of 30 °C and above (Bayliss and Riley 1972a), indicating that chromosome 5D may also be associated with high temperature tolerance. However, this suggestion was based on the scoring of only a few cells because exposure of the plants to high temperatures for 3 days made the chromosomes too sticky to score accurately. Grain number is also reduced much more in N5DT5B plants than in the wild type after exposure to 30 °C during premeiosis and leptotene (Draeger and Moore 2017).
The main aim of this study was to define Ltp1 to a small enough region on 5DL to enable the identification of a candidate gene for the low temperature pairing phenotype. The strategy for mapping Ltp1 was similar to the one used to map the Ph1 locus (Roberts et al. 1999; Griffiths et al. 2006) a major locus that promotes homolog synapsis and regulates crossover formation in wheat (Martín et al. 2014, 2017). This had involved screening for gamma irradiation-induced deletions on specific chromosomes in large populations of wheat. The work on Ph1 established the dose of gamma irradiation likely to give a good rate of deletion discovery. This strategy was applied in the current study to identify deletions of chromosome 5DL using chromosome-specific markers. Recent development of resources such as the Chinese Spring IWGSC RefSeq v1.0 genome assembly (International Wheat Genome Sequencing Consortium (IWGSC) 2018), the Wheat 820 K Axiom® Breeders’ Array probe set (Winfield et al. 2016) and the Ensembl Plants database (Bolser et al. 2016), facilitated the processes of mapping and candidate gene identification. Following the identification of a candidate gene for the low-temperature phenotype, additional experiments were carried out to determine whether tolerance to both high and low temperatures could be controlled by the same locus.
Materials and methods
Plant materials
Deletion lines were generated in bread wheat (Triticum aestivum L., 2n = 6x = 42) var. ‘Chinese Spring’ by gamma irradiation of 1000 seeds at the International Atomic Energy Agency, Vienna (500 seeds irradiated at 250 Gy and 500 at 300 Gy). M1 deletion lines were self-fertilised. M2 lines were genotyped using Kompetitive allele-specific PCR (KASP) to identify plants with interstitial deletions of 5DL. These deletion mutants were exposed to 13 °C for 7 days to identify plants with asynaptic chromosomes at metaphase I of meiosis. Two other Chinese Spring-derived genotypes were used as controls for the KASP genotyping and in the temperature treatment experiments, the standard euploid (wild type) form, (AABBDD) and the nullisomic 5D-tetrasomic 5B (N5DT5B) genotype, which lacks chromosome 5D. Chinese Spring lines with homozygous terminal deletions of chromosome 5DL were obtained from TR Endo, Kyoto University, Japan. These were 5DL-1, 5DL-2, 5DL-5, 5DL-9 and 5DL-13 (Endo and Gill 1996).
DNA extractions
Plants were initially grown in modular trays in a controlled environment room (CER) at 20 °C (day) and 15 °C (night) with a 16-h photoperiod (lights on between 10:00 and 02:00) and 70% humidity. Wheat seedlings were grown to the 2–3 leaf stage, and approximately 5 cm of leaf material was harvested into 1.2-ml collection tubes containing 3-mm tungsten-carbide beads in a 96-well format on dry ice. DNA was extracted using a method based on the protocol available at https://www.ars.usda.gov/ARSUserFiles/60701500/SmallGrainsGenotypingLaboratory/DNAextractionprotocol_ERSGGL.pdf (original reference in Pallotta et al. 2003) except that leaf material was ground in a Geno/Grinder (Spex) at 1500 rpm for 2 min. Extracted DNA was diluted with dH20 so that final DNA template concentrations were between 15 and 30 ng.
Genotyping 5DL terminal deletion lines
Nine chromosome 5D-specific microsatellite markers (Somers et al. 2004), from the GrainGenes database https://wheat.pw.usda.gov/GG3/, were used to map the breakpoints of the five Chinese Spring 5DL terminal deletion lines (Fig. 1). Following PCR amplification, products were separated by agarose gel electrophoresis.
Map of wheat chromosome arm 5DL showing locations of deletions detected using 5DL-specific KASP markers (black or red text with BA prefix) or microsatellite markers (blue); markers are aligned with positions on the Chinese Spring IWGSC RefSeq v1.0 genome assembly (shown in Mb); yellow boxes mark the extent of the 5DL terminal deletions in lines with normal pairing at 13 °C; breakpoints of terminal deletion lines are indicated by blue arrows; pink boxes show positions of 5DL interstitial deletions in lines with normal pairing at 13 °C; orange boxes show the location of the 5DL deletion in the mutant line 22-F5 (ttmei1), which exhibits asynapsis at 13 °C; small grey box shows the ttmei1 deletion region and its flanking markers (in red) as detected in the initial KASP genotyping analysis with the only marker deleted in ttmei1 (BA00822801) shown in bold text; larger grey inset box shows fine mapping between flanking markers BA00334971 and BA00808441 (red text) using BA00822801 and 25 additional KASP markers. Markers shown in orange boxes are deleted in the ttmei1 mutant. The position of the candidate gene TaDmc1-D1 (green box) at 225 Mb is indicated with a black arrow
KASP genotyping of M2 plants
KASP genotyping of M2 plants was performed using SNP (single nucleotide polymorphism) markers from the Wheat Breeders’ 820 K Axiom® array (Winfield et al. 2016) available at www.cerealsdb.uk.net. This data set was aligned with the Chinese Spring reference sequence assembly, IWGSC RefSeq v1.0 (IWGSC 2018), using Python Data Matcher https://github.com/wingenl/python_data_matcher, which merges files with common features. The BioMart data mining tool from Ensembl Plants (Kinsella et al. 2011), available at http://plants.ensembl.org/biomart/martview/, was used as a source for the RefSeq v1.0 assembly coordinates.
Initially, KASP markers used for genotyping were selected at 15-Mb intervals along chromosome 5DL, between the centromere and the breakpoint of the most proximal Chinese Spring terminal deletion line. Later, more KASP markers were used to increase the density across regions of interest (Fig. 1). Selected KASP Primers were 5D chromosome specific and had homeologous SNPs at the 3′ end. The allele-specific forward primers and common reverse primers were synthesised by Sigma-Aldrich. Allele-specific primers were synthesised with standard FAM or VIC compatible tails at their 5′ ends (FAM tail: 5′ GAAGGTGACCAAGTTCATGCT 3′; VIC tail: 5′ GAAGGTCGGAGTCAACGGATT 3′). Twenty primer sets were used in the first round of screening but following the identification of the ttmei1 5DL deletion mutant, 25 more KASP markers were selected between markers BA00334971 and BA00808441 to fine map the deletion (Fig. 1).
KASP reaction and PCR conditions
The KASP reaction and its components were as recommended by LGC Genomics Ltd and described at https://www.biosearchtech.com/support/education/kasp-genotyping-reagents/how-does-kasp-work.
Assays were set up as 5 μl reactions in a 384-well format and included 2.5 μl genomic DNA template (15–30 ng of DNA), 2.5 μl of KASP 2 × Master Mix (LGC Genomics), and 0.07 μl primer mix. Primer mix consisted of 12 μl of each tailed primer (100 μM), 30 μl common primer (100 μM) and 46 μl dH2O. PCR amplification was performed using an Eppendorf Mastercycler Pro 384 thermal cycler (Eppendorf, UK) using the following programme: hot start at 94 °C for 15 min, followed by ten touchdown cycles (94 °C for 20 s; touchdown from 65 to 57 °C for 1 min, decreasing by 0.8 °C per cycle) and then 30 cycles of amplification (94 °C for 20 s; 57 °C for 1 min). Fluorescent signals from PCR products were read in a PHERAstar microplate reader (BMG LABTECH Ltd.). If tight genotyping clusters were not obtained, an additional 5 cycles (94 °C for 20 s; 57 °C for 1 min) were performed. Genotyping data were analysed using KlusterCaller software (LGC Genomics).
Low- and high-temperature treatments
Plants were initially grown in a CER at 20 °C (day) and 15 °C (night) with a 16-h photoperiod (lights on between 10:00 and 02:00) and 70% humidity until Zadoks growth stage 39 (Zadoks et al. 1974; Tottman 1987) when the flag leaf ligule is just visible. They were then transferred to plant growth cabinets under continuous light and exposed to low temperatures (13 °C) for 7 days (with 70% humidity) or high temperatures (30 °C) for 24 h (75% humidity). At 20 °C, meiosis takes around 24 h to complete (Bennett et al. 1971, 1973). However, high temperatures speed up meiosis and low temperatures slow it down (Bennett et al. 1972), so the different lengths of the high- and low-temperature treatments ensured that plants would be exposed to temperature treatments during the period from premeiotic interphase to early meiosis I, when they are most sensitive to temperature stress. It also ensured that the anthers to be sampled would have pollen mother cells (PMCs) containing chromosomes at metaphase I at the end of the treatment period so that synapsis and CO formation could be scored. At 30 °C, 24 h is long enough to ensure meiosis progresses to metaphase I but minimises the time available for adverse effects of high temperature on the plants. Longer periods of high-temperature treatment would have made scoring PMCs more difficult as the chromosomes become sticky (Bayliss and Riley 1972a). To prevent dehydration during high-temperature treatment, plant pots were kept in trays of water. For the high-temperature treatments, plants were placed into the treatment cabinets at approximately the same time of day, between 10.00 and 11.45 am.
Preparation of PMCs for phenotyping
After temperature treatment, anthers were collected from spikes estimated to be undergoing meiosis (when the flag leaf had fully emerged, and the spike length was 4–6 cm long). Anthers were sampled from the first (oldest) 5 tillers only. Using an M80 stereo microscope (Leica Microsystems Ltd., Milton Keynes, UK), anthers were dissected from the two largest florets in each spikelet. Only anthers at metaphase I were scored, so, to determine the meiotic stage, one anther from each floret was stained with acetocarmine and squashed under a cover slip to extrude the PMCs, which were then examined using a DM2000 light microscope (Leica Microsystems). The three anthers within any floret are synchronised in meiotic development, so when PMCs with metaphase I chromosomes were identified, the two remaining anthers from the same floret were fixed in 3:1 (v/v) 100% ethanol/acetic acid, for cytological analysis. Anthers were incubated for at least 24 h at 4 °C before being transferred to 70% ethanol. Fixed anthers were washed with 0.1% sodium dodecyl sulphate for 3–5 min and then hydrolysed with 1 M hydrochloric acid for 10 min at 60 °C. They were then Feulgen-stained with Schiff’s reagent and squashed in 45% acetic acid. This allowed the chromosomes to be spread more widely to facilitate scoring of crossover. Images of metaphase I chromosomes were captured using a DM2000 microscope equipped with a DFC450 camera and controlled by LAS v4.4 system software (Leica Microsystems). For each cell, images were captured in up to 8 different focal planes to aid scoring.
Cytological analysis of chromosome crossover
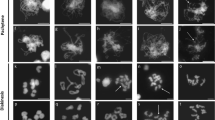
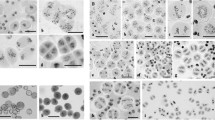
For each plant, 20–30 PMCs were blind scored from digital images. For each cell, the different meiotic chromosome configurations were counted. These were unpaired univalents (0 chiasmata), rod bivalents (1 chiasma), ring bivalents (2 chiasmata), trivalents (2–3 chiasmata), tetravalents (3 chiasmata) and pentavalents (4 chiasmata). Chiasma frequency per PMC was calculated separately for single and double chiasmata (see Fig. 2 for examples of the scored structures).
Feulgen-stained metaphase I chromosomes from PMCs of wild-type Chinese Spring, 22-F5 (ttmei1) mutant and N5DT5B plants treated at different temperatures. a wild type, bttmei1 and c N5DT5B at normal temperatures; d wild type, ettmei1 and f N5DT5B after 7 days at 13 °C; g wild type and h and ittmei1 after 24 h at 30 °C. Examples of univalent chromosomes (univ), rod bivalents (rod), ring bivalents (ring), multivalents (multiv), single chiasma (X), double chiasmata (XX) and sticky, clumping chromosomes (clump) are indicated with arrows; note complete univalence in ttmei1 and N5DT5B after treatment at 13 °C. Scale bars, 10 μm
Statistical analyses were performed using STATISTIX 10.0 software (Analytical Software, Tallahassee, FL, USA). All treatments were analysed by the Kruskal–Wallis test (nonparametric one-way analysis of variance). Means were separated using the Dunn’s test with a probability level of 0.05. Statistical analysis was carried out between temperatures and between genotypes. Bar charts were plotted using Microsoft Excel (2016).
Immunolocalisation of meiotic proteins ASY1 and ZYP1
To determine when meiosis is disrupted in the low-temperature experiments, PMCs from three wild-type and three ttmei1 mutant plants were immunolabelled with antibodies against the meiotic proteins ASY1 and ZYP1. PMCs from wild-type Chinese Spring and the ttmei1 mutant were embedded in acrylamide pads to preserve their 3D structure, and immunolocalisation of ASY1 and ZYP1 was performed as described previously (Martín et al. 2014, 2017). Anti-TaASY1 (Boden et al. 2009) raised in rabbit was used at a dilution of 1:250, and anti-HvZYP1 (Colas et al. 2016) raised in rat was used at a dilution of 1:200. Anti-rabbit Alexa Fluor® 488 and anti-rat Alexa Fluor® 568 (Invitrogen) were used as secondary antibodies.
Image acquisition and analysis
Images were taken of 30 PMCs from each of the Chinese Spring wild-type and ttmei1 replicates. Polyacrylamide-embedded PMCs were optically sectioned using a DM5500B microscope (Leica Microsystems) equipped with a Hamamatsu ORCA-FLASH4.0 camera and controlled by Leica LAS-X software v2.0. Z-stacks were deconvolved using Leica LAS-X software. Images were processed using Fiji, which is an implementation of ImageJ, a public domain program available from http://rsb.info.nih.gov/ij/ (Schneider et al. 2012).
Dmc1 sequence analysis
Multiple sequence alignment was carried out using ClustalX2 software (Thompson et al. 1997; Larkin et al. 2007). BLAST searches of the Dmc1 sequences from other plant species against the Chinese Spring IWGSC RefSeq v1.0 sequence assembly (IWGSC 2018) revealed that there are three homeologs of the Dmc1 gene in hexaploid wheat: TraesCS5A02G133000 (TaDmc1-A1) on chromosome 5A, TraesCS5B02G131900 (TaDmc1-B1) on 5B and TraesCS5D02G141200 (TaDmc1-D1) on 5D. The nucleotide sequences of the DNA, the coding sequences (CDs) and the promoter regions (including 1500 nucleotides downstream of the start codon) of the three TaDmc1 homeologs were compared.
Multiple DMC1 amino-acid sequence alignment was also carried out between hexaploid wheat (Chinese Spring), barley (Hordeum vulgare) and rice (Oryza sativa Japonica). Gene IDs from Ensembl Plants are, for barley: HORVU5Hr1G040730.3 (HvDMC1) and for rice: Os12g0143800 (OsDMC1A) and Os11g0146800 (OsDMC1B).
The Dmc1 sequences from hexaploid wheat (T. aestivum) were also compared with those of its diploid and tetraploid progenitors, T. urartu (the A-genome donor), Aegilops tauschii (the D-genome donor) and T. dicoccoides (the AABB tetraploid progenitor). Gene IDs from Ensembl Plants are for T. urartu (TuDMC1): TRIUR3_13472; for Ae. tauschii (AetDMC1-D): AET5Gv20357200; for the T. dicoccoides A copy (TdDMC1-A): TRIDC5AG022500 and for the T. dicoccoides B copy (TdDMC1-B): TRIDC5BG023380.
KASP genotyping of M3 plants
KASP genotyping of Chinese Spring wild type and ttmei1 M3 mutant plants was carried out to assess whether the gamma irradiation of Chinese Spring seeds had resulted in a duplication or insertion of Dmc1-D1 in other parts of the genome. Two different sets of Dmc1-D1 chromosome-specific primers were used to genotype three wild type and nine ttmei1 mutant plants. Primer set 1 consisted of two allele-specific primers: 5′ GAAGGTCGGAGTCAACGGATTcctgtcatgaaaccctgactC 3′ (including HEX tail [wild type]) and 5′ GAAGGTGACCAAGTTCATGCTcctgtcatgaaaccctgactT 3′ (including FAM tail [mutant]), and a common reverse primer: 5′ CcatctgtgctatGcgatAgaA 3′. Primer set 2 consisted of allele-specific primers: 5′ GAAGGTCGGAGTCAACGGATTgtagcttagctcctaaacctCaC 3′ (with HEX tail [wild type]) and 5′ GAAGGTGACCAAGTTCATGCTgtagcttagctcctaaacctCaT 3′ (with FAM tail [mutant]), and common reverse primer: 5′ GcgatAgaAtcttctgaaGtttgtG 3′.
Results
Phenotyping 5DL terminal deletion lines at low temperatures
To locate the gene(s) responsible for the Ltp1 phenotype, we first needed to narrow down our region of interest on chromosome 5DL. To facilitate this, we exposed five Chinese Spring lines with homozygous terminal deletions of 5DL to 13 °C for 7 days. All five had normal chromosome pairing at metaphase I. The breakpoints of these deletion lines were mapped using nine 5D-specific microsatellite markers (Fig. 1). The mapping results were consistent with the order of the breakpoints on the C-banding maps produced by Endo and Gill (1996). Line 5DL-1 had the largest terminal deletion. The breakpoint of this deletion lies between the markers Xcfd40 and Xgdm138, the latter being the most proximal marker to be deleted in this line. We inferred from this that the gene responsible for the Ltp1 phenotype must lie proximal to Xgdm138. BLAST searches for the Xgdm138 primer sequence against the Chinese Spring IWGSC RefSeq v1.0 sequence assembly revealed its location to be at approximately 391 Mb. The length of chromosome 5D is estimated to be around 565 Mb and the centromere lies at 185.6–188.7 Mb (IWGSC 2018), so we can infer that the length of 5DL is around 376 Mb. The length of the chromosome distal to Xgdm138 is 174 Mb, so almost half of the chromosome arm could be eliminated from our search to find the gene responsible for the Ltp1 phenotype.
Identification of a 5DL deletion line with abnormal crossover frequency
To generate deletion lines, 1000 Chinese Spring seeds were gamma irradiated at either 250 or 300 Gy. Of the 500 seeds irradiated at 250 Gy, 409 plants (82%) germinated and 378 plants (76%) were fertile. Of the 500 seeds irradiated at 300 Gy, 417 plants (83%) germinated and 372 (74%) were fertile. M1 plants were self-fertilised (four ears per plant where possible), and 1–2 M2 seeds from each ear were genotyped. A total of 2444 Chinese Spring M2 plants were genotyped, initially using 20 KASP markers located in the proximal half of 5DL. This identified 16 plants with deletions in 5DL (Fig. 1). These deletion lines were all derived from seed irradiated with the higher dose (300 Gy) of gamma irradiation. The largest of these deletions (in the mutant line 21-D11) was estimated to be between 16 and 23 Mb.
All 16 deletion mutants were exposed to 13 °C for 7 days during premeiotic interphase to early meiosis I. Fifteen plants had normal bivalent formation at metaphase I, but one deletion mutant, 22-F5, exhibited almost complete crossover failure after low-temperature treatment (Fig. 2e; Table 1; Fig. 3, Supplementary Table 1). This is a very similar phenotype to that described for the Ltp1 locus, so it suggests that Ltp1 is amongst the genes that have been deleted in the 22-F5 mutant.
Bar charts showing genotypic effects on meiotic metaphase I chromosomes of Chinese Spring (CS) wild-type and 22-F5 (ttmei1) mutant plants after treatment at 20 °C, 13 °C and 30 °C. The numbers of univalents, ring and rod bivalents and single crossovers are shown. Multivalents and double crossovers are not shown
Given the effects of low temperature on the 22-F5 mutant, we assessed whether it also showed reduced tolerance to high temperature. After 24 h at 30 °C, the 22-F5 deletion mutant did indeed exhibit a reduced number of crossovers and increased univalence, but to a lesser extent than at 13 °C (Table 1; Fig. 3; Suppl. Table 1). Therefore, we renamed the 22-F5 deletion mutant ‘ttmei1’ to reflect the fact that it has a deletion of an unknown gene ‘TTmei1’ (Temperature-Tolerant meiosis 1).
Crossover in ttmei1 mutants at normal temperatures
The ttmei1 mutant plant was self-fertilised and was able to produce M3 seed, although seed numbers appeared to be lower than that produced by wild-type plants. Further phenotypic analysis was carried out on Chinese Spring wild type and ttmei1 M3 plants, which were grown under normal temperature conditions or were exposed to 13 °C for 7 days. Five plants were used for each treatment group, and 20–30 PMCs were scored for each plant. Statistical analysis identified differences in synapsis and crossover at different temperatures and between the two different genotypes.
At 20 °C, in wild-type plants, all chromosomes lined up on the metaphase plate as normal, pairing as bivalents, mostly as rings but with the occasional rod bivalent (Fig. 2a). However, at the same temperature, the numbers of univalents, rod bivalents and multivalents were significantly higher in the ttmei1 mutant when compared with the wild type, and the numbers of ring bivalents and both single and double crossovers were significantly lower (Table 1). Although the proportion of ring and rod bivalents was significantly different in ttmei1, the mean number of bivalents (rings + rods) was around 19 as opposed to 21 in the wild type, and the chromosomes were still able to align on the metaphase plate with the exception of the occasional one or two univalent chromosomes (Fig. 2b). At 20 °C, ttmei1 chromosomes had similar conformations to those observed in N5DT5B plants (Fig. 2c).
Crossover failure under low temperatures in ttmei1 mutants
The 13 °C treatment had no significant effect on wild-type metaphase I chromosomes (Table 2), which paired mainly as ring bivalents and lined up on the metaphase plate as for normal temperatures (Fig. 2a, d). However, in the ttmei1 mutant, there were significantly higher numbers of univalent chromosomes at 13 °C than at 20 °C and significantly lower numbers of ring bivalents and crossovers (Table 2; Fig. 4). In ttmei1 mutant plants at 13 °C, the mean number of univalents per cell was almost 40, whereas in wild-type plants at the same temperature the mean number of univalents was less than one (Table 1; Fig. 3). In fact, in 60% of ttmei1 PMCs treated at 13 °C, all chromosomes were univalent, with 24–41 univalent chromosomes in the remaining 40% of PMCs (data not shown). Univalents appeared more condensed than other chromosomes and were unable to align on the metaphase plate (Fig. 2e). The low chiasma frequency and high numbers of univalent chromosomes in ttmei1 at low temperatures are very similar in effect to that described when the whole of chromosome 5D is deleted in N5DT5B, as shown in Fig. 2f.
Bar charts showing the effects of three different temperature treatments (20 °C, 13 °C and 30 °C) on meiotic metaphase I chromosomes of Chinese Spring (CS) wild-type and 22-F5 (ttmei1) mutant plants. The numbers of univalents, ring and rod bivalents and single crossovers are shown. Multivalents and double crossovers are not shown
Immunolabelling of PMCs at low temperatures
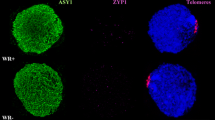
To investigate when meiosis was being disrupted in the low-temperature experiments, wild-type and ttmei1 mutant PMCs were immunolabelled with antibodies against the meiotic proteins ASY1 and ZYP1 to follow the progression of synapsis (Fig. 5). ASY1 is part of the lateral elements of the SC and first appears during premeiotic interphase, before synapsis begins (Armstrong et al. 2002; Boden et al. 2009). ZYP1 is part of the central region of the SC, which assembles between the lateral elements (Higgins et al. 2005; Khoo et al. 2012), and it is present only where chromosomes are synapsed. Immunolabelling therefore gives an indication of the level of synapsis achieved at pachytene when the SC is fully established, which enables tracking of the progression of synapsis. Clear differences were detected when comparing ASY1 (green) and ZYP1 (magenta) signals in the wild type and ttmei1 mutant at low temperatures. In wild-type wheat (Fig. 5a), synapsis proceeds in the same way as it does at normal temperatures: It starts from the telomeres at one pole of the nucleus during early zygotene, and, by pachytene, all chromosomes have synapsed. A very small amount of ASY1 labelling is still visible at pachytene, indicating the small amount of chromatin that has not yet synapsed. In ttmei1 mutants (Fig. 5b), at early zygotene, ASY1 localises to the chromosome axes in a pattern similar to that observed in wild-type wheat, and synapsis initiates normally at one pole of the nucleus. After early zygotene, chromatin staining with DAPI (4′,6-diamidino-2-phenylindole) suggests that the ttmei1 PMCs are at pachytene (Fig. 5b), but the amount of ZYP1 labelling is much lower than in pachytene in the wild type, indicating that synapsis has been compromised and is not completed. None of the PMCs had a completed synapsis in the ttmei1 mutants.
Immunolocalisation of meiotic proteins ASY1 (green) and ZYP1 (magenta) in PMCs from wheat Chinese Spring wild type (a) and ttmei1 mutant (b), both under low-temperature conditions. a During early zygotene, synapsis starts from the telomeres at one pole of the nucleus in wild-type wheat. During pachytene, all chromosomes have synapsed. ASY1 labelling in green shows that a very small proportion of chromatin has not yet synapsed. b In ttmei1 mutants, synapsis initiates normally at one pole of the nucleus; however, synapsis is soon compromised and is not completed. Therefore, there is no normal pachytene in the ttmei1 mutant. DAPI staining in blue. Scale bar, 10 μm
Gene content in the ttmei1 deletion region
The ttmei1 deletion was initially mapped to a 16-Mb region of 5DL, between the flanking markers BA00334971 and BA00808441 (Fig. 1). In this initial analysis, only one KASP marker, BA00822801, was deleted in ttmei1. KASP genotyping of ttmei1 using 25 more markers between BA00334971 and BA00808441 fine-mapped the deletion to a 4-Mb region between BA00798994 and BA00124919 (Fig. 1). Seven markers were deleted in this region.
The gene content within the 4-Mb deletion regions in ttmei1 was revealed using data derived from the hexaploid wheat gene annotation v1.1 (IWGSC 2018) available from Ensembl Plants. Functional annotations of the genes were retrieved from the file ‘FunctionalAnnotation.rds’ in https://opendata.earlham.ac.uk/wheat/under_license/toronto/Ramirez-Gonzalez_etal_2018-06025-Transcriptome-Landscape/data/TablesForExploration/FunctionalAnnotation.rds (Ramírez-González et al. 2018). Expression patterns of all the genes within the ttmei1 deletion region were investigated based on 876 RNASeq samples from different tissue types available in the wheat expression browser http://www.wheat-expression.com. A total of 41 genes (16 high confidence and 25 low confidence genes) were identified in the 4-Mb deletion region. Eighteen of the 41 genes were expressed during meiosis (Table 3). We considered a gene to be expressed when its expression level was > 0.5 transcripts per million (TPM) in at least one meiotic sample (Alabdullah et al. 2019). Twelve of these eighteen genes are high confidence genes. Detailed expression data (TPM) for all genes in the ttmei1 deletion region are shown in Supplementary Table 2.
TaDmc1-D1 is a candidate for the Ltp1 phenotype
Of the 18 genes expressed during meiosis in the ttmei1 deletion interval, the strongest candidate for the TTmei1 phenotype is TraesCS5D02G141200, which is the D-genome homeolog of Dmc1 (Disrupted meiotic cDNA 1), which has a known meiotic phenotype, playing a central role in homologous recombination. It is a high confidence gene, and its expression level is around ten times higher in meiotically active tissues compared with non-meiotic tissues (averages 26.9 TPM and 2.8 TPM, respectively [Suppl. Fig. 2]). The other 17 genes in the region are much less likely than TaDmc1 to be candidates for the TTmei1 phenotype because none of them could be attributed to any known meiotic function, and although they are expressed during meiosis, expression is proportionally higher in other tissues.
To assess whether gamma irradiation of the Chinese Spring seeds had resulted in a duplication or insertion of Dmc1-D1 in other parts of the genome, three Chinese Spring wild-type and nine ttmei1 M3 mutant plants were KASP-genotyped using two different sets of Dmc1-D1-specific primers. As expected, with both primer sets, none of the ttmei1 mutant plants showed any amplification when compared with the wild-type plants. This indicates very clearly that Dmc1-D1 is completely deleted from the ttmei1 deletion line and has not been duplicated or inserted elsewhere in the genome as a result of the gamma irradiation.
BLAST searches against the Chinese Spring IWGSC RefSeq v1.0 sequence assembly (IWGSC 2018) revealed that there are three homeologs of the Dmc1 gene in hexaploid wheat: TraesCS5A02G133000 (TaDmc1-A1) on chromosome 5A, TraesCS5B02G131900 (TaDmc1-B1) on 5B and TraesCS5D02G141200 (TaDmc1-D1) on 5D. There are differences in gene expression levels between the three Dmc1 homeologs in meiotic anthers: TaDmc1-A1 has the lowest gene expression levels and TaDmc1-D1 the highest (Suppl. Fig. 1).
Dmc1 sequence analysis
The nucleotide sequences of the DNA, the coding sequences (CDs) and the promoter regions of the three TaDmc1 homeologs were compared. The CDs of the A, B and D homeologs are highly conserved (Suppl. Fig. 2) with sequence identity ranging between 98.1 and 98.7% (Suppl. Table 3a). The DNA sequences are less similar (Suppl. Fig. 3), with sequence identity ranging from 82.1% (between A and B homeologs) to 89.0% (between A and D homeologs) (Suppl. Table 3a). Comparing the promoter regions of each of the three TaDmc1 homeologs showed a large insertion mutation (163 nt in size) at position 430 downstream of the start codon of TaDmc1-B1 (Suppl. Fig. 4).
The exon–intron structure of the three TaDmc1 homeologs (Fig. 6a) is conserved in terms of number of exons (14 exons each); however, there is a variation in intron length between the three homeologs due to large indel mutations mainly in introns 3, 6 and 7 (data not shown). TaDmc1 homeologs have no splice variants. In all three TaDmc1 homeologs, there is an open reading frame of 1035 bp (Suppl. Fig. 2), which encodes a predicted protein of 344 amino acids (Fig. 6b). This protein is highly conserved between the three homeologs. There are five single amino-acid substitutions between the wheat homeologs, but only one single amino-acid substitution (at position 114) in TaDMC1-D1 (threonine) when compared with TaDMC1-A1 and TaDMC1-B1 (alanine) (Fig. 6b).
TaDmc1 gene structure and amino-acid sequence alignment. a Exon–intron structure of the three TaDmc1 homeologs. b Multiple sequence alignment of amino acids from DMC1 proteins of different cereal plants: hexaploid wheat (Triticum aestivum cv Chinese Spring; Ta), barley (Hordeum vulgare; Hv) and rice (Oryza sativa Japonica; Os). SNPs are shown in red text on a grey background. Symbols below each position in the sequence indicate the amount of conservation (asterisk ‘*’: identical residues; colon ‘:’: conserved substitution; period ‘.’: semi-conserved substitution; and space ‘ ’: not conserved). A single amino-acid substitution in TaDMC1-D1 at position 114 (indicated by a large red triangle) may confer low temperature tolerance in wheat. Small blue triangles show the positions of three other amino-acid substitutions in TaDMC1-B1 at positions 167, 214 and 316, and a small green triangle shows the position of a substitution in TaDMC1-A1 at position 310
Multiple DMC1 amino-acid sequence alignment was also carried out between the three hexaploid wheat homeologs and barley (Hordeum vulgare) and rice (Oryza sativa Japonica). There is a high level of amino-acid sequence conservation between the wheat, barley and rice proteins (Fig. 6b) with sequence identity ranging between 95.3 and 99.4% (Suppl. Table 3b). This conservation of amino-acid sequence between the wheat, barley and rice DMC1 proteins has also been reported by Colas et al. (2019).
Dmc1 sequence comparisons were also made between hexaploid wheat (T. aestivum) and its diploid and tetraploid progenitors, T. urartu (the A-genome donor), Ae. tauschii (the D-genome donor) and T. dicoccoides (the AABB tetraploid progenitor) (Suppl. Fig. 5). The Dmc1 sequences of T. aestivum are quite conserved when comparing each gene copy with its ancestral genome copy. For the DNA sequences, the highest percentage of homology is between the D copy of T. aestivum and the ancestral gene of Ae. tauschii (99.8%), whilst the lowest homology is between the B copy of T. aestivum and the B-genome copy in T. dicoccoides (92.2%) (Suppl. Table 3c), due to a large insertion of 294 bp in intron 7 (Suppl. Fig. 5, Suppl. Fig. 6). The percentages of homology in TaDmc1-A1 versus T. dicoccoides_A and TaDmc1-A1 versus T. urartu are 98.6% and 98.4%, respectively (Suppl. Table 3c).
Comparisons were also made between the amino-acid sequences of DMC1 proteins from T. aestivum and those of its diploid and tetraploid progenitors (Suppl. Fig. 7). Unlike those of T. aestivum, most of the Dmc1 gene copies in the wheat ancestors have multiple transcripts due to alternative splicing variants (as predicted by Ensembl Plants). TdDmc1-A (T. dicoccoides A copy) has 3 transcripts; TdDmc1-B (T. dicoccoides B copy) has 9 transcripts; and AetDmc1-D (Ae. tauschii) has 16 transcripts. TuDmc1-A (T. urartu, TRIUR3_13472) has a single transcript, but of all the ancestral DMC1 proteins in our comparisons, the encoded protein sequence of TuDMC1 from T. urartu is the one that diverges most from that of T. aestivum (Suppl. Table 3d). Of the amino-acid differences between the A-, B- and D-genome copies of DMC1 in T. aestivum, the single amino-acid substitution in the D-genome copy of T. aestivum at position 114 (Fig. 6) is also present in the D-genome ancestor, and the A-genome substitution at position 310 is also present in the A-genome ancestor, whereas the B-genome substitutions at positions 167, 214 and 316 are different to all of the ancestral copies in T. dicoccoides (Suppl. Fig. 7).
Crossover failure under high temperatures in ttmei1 mutants
Five wild-type and 11 ttmei1 M3 plants were exposed to 30 °C for 24 h. A higher number of mutant plants were treated because of the wide variation in the data for this group. Chromosome stickiness and clumping were observed in many of the cells at this temperature (Fig. 2i), which made scoring more difficult than at normal or low temperatures. In wild-type plants, high temperature resulted in significantly higher numbers of rod bivalents and significantly lower numbers of ring bivalents and crossovers than had been observed at 20 °C (Figs. 2g, 4; Table 2). However, these differences were much more pronounced in the ttmei1 mutant. In ttmei1 mutant plants, there were significantly higher numbers of univalents, rod bivalents and tetravalents at 30 °C than at normal temperatures and significantly fewer ring bivalents and crossovers although the level of univalence was not nearly as pronounced as that at 13 °C (Table 2; Fig. 4). There were also significant differences between the two genotypes at 30 °C. For example, the mean number of univalents was 7.85 in ttmei1 mutants but only 0.21 in wild-type plants (Table 1; Fig. 3). It was also observed that many ttmei1 chromosomes did not align correctly on the metaphase plate (Fig. 2h, i).
Discussion
Chinese Spring wild-type plants and mutant plants with interstitial deletions within chromosome 5DL were exposed to low temperature (13 °C) for 7 days during a period lasting from premeiotic interphase to early meiosis I. Microscopic examination of PMCs from these plants enabled us to identify a 5DL deletion mutant whose meiotic chromosomes exhibit extremely high levels of asynapsis and chromosome univalence. Within the deleted region of this mutant, we identified a candidate gene on 5DL (TaDmc1-D1) that is probably responsible for the Ltp1 phenotype. Subsequent exposure of this ltp1 deletion mutant to 24 h of high temperature (30 °C) during the same developmental period led to a reduced number of crossovers and increased univalence, though to a lesser extent than at 13 °C. As this deletion clearly has an effect on chromosome pairing at 30 °C as well as at 13 °C, the mutant line was renamed ttmei1 to reflect both its reduced high temperature tolerance and its loss of Ltp1.
By exploiting a series of wheat 5DL terminal deletion lines, the Ltp1 locus was initially delimited to the proximal half of chromosome 5DL. Using terminal deletion lines to narrow down our region of interest proved to be a good strategy as we were able to eliminate almost the entire distal half of the chromosome from our investigations. The deletion mapping was refined by screening a population of around 2500 gamma-irradiated plants using 5DL-specific KASP markers. Of these, sixteen plants had 5DL deletions. The resources now available, including the Chinese Spring RefSeq v1.0 genomic sequence assembly, KASP primers from www.cerealsdb.uk.net and the Ensembl Plants database, made the screening and mapping processes much easier and quicker than when a similar approach was used to map the Ph1 locus (Griffiths et al. 2006).
Exposure of the 5DL mutant plants to 13 °C for 7 days identified a deletion mutant, ttmei1, that exhibits extremely high levels of chromosome univalence at low temperatures. This is very similar to the Ltp1 phenotype previously described in N5DT5B plants where the whole of chromosome 5D is missing. When grown under normal conditions, ttmei1 plants are not sterile and are able to produce seed, however, seed numbers appeared to be lower than in the wild type. In ttmei1 mutants, chromosome synapsis and crossover are only slightly disrupted at 20 °C, but multivalents were observed in four of the ttmei1 mutants at this temperature, which could indicate the presence of additional rearrangements that might alter chromosome pairing. However, in one mutant plant, ttmei1-4, no multivalents were observed (Suppl. Table 1), suggesting that the ttmei1 deletion is not necessarily linked to the occurrence of multivalents. The presence of high numbers of univalents and more rod bivalents than normal in ttmei1 mutants under low temperatures suggests a major problem with CO formation. Consistent with this, the ttmei1 line exhibits significant abnormalities of synapsis at low temperatures.
The ttmei1 mutant carries a 4-Mb deletion of 5DL. Within this deletion interval, there are 41 genes, 18 of which are expressed during meiosis. Amongst these genes, the strongest candidate for the low-temperature pairing phenotype is the meiotic recombination gene TaDmc1-D1. None of the other genes are known to have any meiotic function. Moreover, TaDmc1 is expressed most highly during early prophase I (Suppl. Table 2), which, in wheat, is when synapsis is initiated at the ‘telomere bouquet’ stage (Martín et al. 2017). TaDmc1-D1 is therefore a strong candidate for temperature tolerance at low temperatures.
DMC1 plays a central role in homologous recombination, SC formation and cell-cycle progression. It is a meiosis-specific protein, structurally similar to the bacterial strand exchange protein RECA (Bishop et al. 1992). The control of DNA strand exchange during meiotic recombination is vital for sexual reproduction. Homologous recombination is initiated by the programmed formation of DNA double-strand breaks (DSBs) at leptotene. DMC1 and RAD51 (another RECA homolog) form filaments on the single-strand DNA overhangs at DSBs. These filaments facilitate homology search and catalyse strand invasion and strand exchange between homologous chromosomes (Neale and Keeney 2006). Repair of these interhomolog invasion events results in crossovers or non-crossovers (Reviewed in Lambing et al. 2017).
Dmc1 homologs are found in a wide variety of organisms. In yeast, dmc1 mutants have defects in reciprocal recombination, fail to form normal SCs, accumulate DSB recombination intermediates and the cells arrest in late meiotic prophase (Bishop et al. 1992). In mice, dmc1 mutants have defective synapsis which leads to severe sterility also due to prophase arrest (Pittman et al. 1998; Yoshida et al. 1998; Bannister et al. 2007). In our wheat experiments, immunolocalisation of ASY1 and ZYP1 showed that at 13 °C synapsis was compromised in ttmei1 mutants and was not completed, with meiosis appearing to arrest before pachytene in late prophase. This is similar to the phenotypes observed in yeast and mice at ambient temperatures.
In most diploid plant species, deletion of Dmc1 also leads to sterility (Devisetty 2010). However, in Arabidopsis thaliana, atdmc1 mutants have a disrupted synapsis with high levels of univalence, resulting in abnormal pollen grain formation and reduced fertility, but plants are not completely sterile because random chromosome segregation allows enough PMCs to reach maturity (Couteau et al. 1999). The Arabidopsis genome contains one copy of the Dmc1 gene. In rice (Oryza sativa), there are two Dmc1 homologs, OsDMC1A and OsDMC1B, that probably arose through chromosome duplication (Ding et al. 2001). A mutation in one or other of these homologs does not cause problems in meiosis, but Osdmc1a Osdmc1b double mutants exhibit serious CO defects, abnormal synapsis, high numbers of univalents at metaphase and are sterile (Wang et al. 2016). Barley (Hordeum vulgare) carries a single Dmc1 homolog, HvDMC1, and mutations in this gene lead to abnormal synapsis, multiple univalents and chromosome mis-segregation (Colas et al. 2019; Szurman-Zubrzycka et al. 2019). Thus, it appears that disruption of the barley orthologue of Dmc1 at normal temperatures leads to a phenotype similar to that of ttmei1 at low temperatures.
Previous evidence from animal studies supports our suggestion that Dmc1 is involved in the maintenance of normal chromosome synapsis at low temperatures. In the Japanese red-bellied newt, Cynops pyrrhogaster, which is a poikilothermic amphibian, low temperatures during meiosis impair synapsis/recombination, which results in the production of abnormal spermatozoa (Yazawa et al. 2003). In newts incubated at 8 °C, spermatogenesis does not proceed beyond meiotic metaphase I, and univalent chromosomes are observed, indicating chromosome pairing failure as a result of asynapsis. At 12 °C, meiotic chromosomes have similar, though less severe, abnormalities. Under the same low-temperature conditions (8 °C or 12 °C), expression of the DMC1 protein decreases, leading to the suggestion that its low expression level contributes to the temperature-dependent abnormalities in spermatogenesis.
In hexaploid wheat, there are three copies of Dmc1: TaDmc1-A1 on 5A, TaDmc1-B1 on 5B and TaDmc1-D1 on 5D, and differences in the abilities of these three genes to stabilise the genome at low temperatures could also be related to differences in gene expression. In the hexaploid wheat variety ‘Highbury’, there are differences in the levels of gene expression between the three Dmc1 homeologs during premeiosis and early meiosis, which suggests that they might contribute to meiotic recombination to differing extents (Devisetty et al. 2010). This is reflected in our own study in Chinese Spring, where, in meiotic anthers, gene expression levels are highest in TaDmc1-D1, intermediate in TaDmc1-B1 and lowest in TaDmc1-A1 (Suppl. Fig. 1). In Chinese Spring, deletion of the region containing TaDmc1-D1 results in asynapsis at 13 °C, and it seems highly likely that this gene promotes low temperature tolerance. However, different dosages of the TaDmc1 alleles can affect the stability of synapsis at low temperatures. When chromosome 5A is present as a single dose in N5DT5B plants, it cannot compensate for the lack of 5D and is unable to stabilise synapsis at low temperatures, but when present as a double dose in N5DT5A plants it can compensate for the 5D deficiency and chromosome pairing at 12 °C is normal (Riley et al. 1966). The A-genome homeolog, TaDmc1-A1, has the lowest expression of the three copies of Dmc1 in Chinese Spring. This suggests that when Dmc1-A1 is present at a double dose, an increase in its expression compensates for the loss of Dmc1-D1 to provide low temperature tolerance. There is only one amino-acid difference between the TaDMC1-A1 and TaDMC1-D1 proteins (Fig. 6b), a substitution of alanine (a non-polar amino acid) in TaDMC1-A1 (and TaDMC1-B1) by threonine (a polar amino acid) in TaDMC1-D1 at position 114. The threonine residue is also present in the ancestral genome of Ae. tauschii (Suppl. Fig. 7). The fact that TaDMC1-A1 can compensate for TaDMC1-D1 suggests that this amino-acid difference is not so important for low temperature tolerance.
The A- and D-genome copies of TaDmc1 are highly conserved with their ancestral homeologs, which suggests that these copies are functional. The B-genome copy is less well conserved due to a large insertion of 294 bp in intron 7 (Suppl. Fig. 5; Suppl. Fig. 6). Dmc1-B1 has intermediate gene expression in Chinese Spring, higher than Dmc1-A1, but lower than Dmc1-D1, but it cannot compensate for the loss of the D copy at low temperatures even when present as a double dose. The inability of Dmc1-B1 to stabilise low-temperature pairing may arise from three amino-acid differences in TaDMC-B1 compared to the A- and D-genome copies, or from the 163-bp insertion in the promoter region of TaDmc1-B1. Alternatively, the 294-bp insertion in intron 7 of TaDmc1-B1 could be affecting gene expression, because although intronic sequences do not encode protein products, they can still have an effect on gene expression and gene functionality (Mattick and Makunin 2006; Rearick et al. 2011). It is important that breeders are aware that there could be TaDmc1 alleles in their wheat germplasm that cannot stabilise wheat synapsis at extremes of temperature and that the effects of these alleles may be masked by the temperature-stabilising actions of TaDMC1-D1.
In tetraploid wheats (AABB), synapsis at 12 °C is normal despite the absence of chromosome 5D (Riley et al. 1966). This may be due to the presence of a dominant Ltp allele on chromosome 5A that duplicates the chromosome stabilising activity of 5D in hexaploid wheat (Hayter and Riley 1967). Interestingly, a previous study has shown that some other varieties and subspecies of wheat differ from Chinese Spring in that the gene that stabilises chromosome pairing at low temperatures is located on a chromosome other than 5D (Chapman and Miller 1981). In the wheat variety ‘Hope’ and the subspecies spelta, the dominant Ltp gene is located on chromosome 5A rather than 5D.
It has also been postulated that there are additional Ltp genes on chromosomes 5AS (Ltp2) and 5BS (Ltp3) that also have a stabilising effect on homologous pairing and crossover at low temperatures (Queiroz et al. 1991), but high levels of univalence and low chiasma frequencies were only seen after 20 days of cold treatment and at a slightly lower temperature (10 °C).
To determine whether TaDmc1-D1 is the gene responsible for stabilising the low-temperature asynapsis observed in Chinese Spring plants with 5D chromosomal deletions, the next step will be to either disrupt or delete the gene and then assess the resulting mutant plants for low-temperature asynapsis. This could potentially be done either using Targeting Induced Local Lesions IN Genomes (TILLING) populations or by CRISPR/Cas9 genome editing. TILLING lines are available in the hexaploid wheat variety ‘Cadenza’ but are not yet available for Chinese Spring. Moreover, results from recent experiments to edit a gene within the Ph1 locus suggest that CRISPR may be a better choice for deleting the TaDmc1-D1 gene. Like Ltp1, the Ph1 locus, on wheat chromosome 5B, is scored by its deletion phenotype. This locus, which affects both chromosome pairing and crossover, contains the major wheat meiotic crossover gene ZIP4. When zip4 CRISPR deletion mutants were compared with the ph1b deletion mutant (Sears 1977), they exhibited a similarly high level of homeologous crossover and a similar effect on homologous pairing (Rey et al. 2018). Whereas, when Cadenza zip4 TILLING mutants were compared to the ph1b mutant, although they exhibited a similarly high level of homeologous crossover, only some exhibited a similar effect on homologous pairing (Rey et al. 2017). Therefore, we are currently investigating the possibility of generating TaDmc1-D1 Chinese Spring deletion mutants using CRISPR/Cas9 genome editing.
In ttmei1 mutants, aberrant chiasma formation was also observed after 24 h at 30 °C, though this was not as pronounced as in the low-temperature phenotype. So, if TaDmc1-D1 is the gene responsible for the observed low temperature tolerance, it may well also have a stabilising effect at high temperatures, though to a lesser extent than at low temperatures. There is high variation in the scores for chiasma frequency between the 11 ttmei1 plants treated at 30 °C. One explanation for this variation could be that the faster progression of meiosis at high temperature may make it harder to target the stages most sensitive to high temperatures. Plants were placed into the 30 °C treatment cabinets at slightly different times of day (10.00–11.45 am), so that high-temperature treatment may have coincided with temperature-sensitive stages of meiosis or premeiosis to a greater or lesser extent.
However, it is also likely that ttemi1 mutants carry many duplications and deletions in addition to those we have identified on 5DL, particularly since the ttmei1 mutant plants were derived from seed irradiated with the higher dose (300 Gy) of gamma irradiation. We have determined through KASP genotyping that Dmc1-D1 has not been duplicated or inserted elsewhere in the genome, but it is possible that ttmei1 mutants carry a background deletion that has not yet been detected that contains a different meiotic recombination gene affecting high temperature tolerance that is segregating with the ttmei1 deletion. However, all 11 of the ttemi1 mutants scored at high temperatures exhibited reduced crossover, suggesting that the ttemi1 deletion is associated with reduced high temperature tolerance. We are currently conducting further experiments to understand this variation at high temperature. We are also backcrossing ttmei1 mutants with Chinese Spring wild-type plants to remove background deletions.
Implications for plant breeding
In the future, breeders face the challenge of producing crops with increasing resilience to environmental stress whilst producing ever higher yields (Steinmeyer et al. 2013). Global climate models predict that temperatures will continue to increase over the twenty-first century and that high-temperature extremes such as heat waves are likely to occur with a higher frequency and duration (Collins et al. 2013). It has been estimated that if global temperatures increase by just one degree (°C), wheat yields will decrease by 6% (Asseng et al. 2015). In the main European wheat-growing areas, the occurrence of adverse weather conditions might substantially increase by 2060, resulting in more frequent crop failure (Trnka et al. 2014). This could threaten global food security since Europe produces almost a third of the world’s wheat.
Moreover, the probability of multiple adverse events occurring within one season is projected to increase sharply by mid-century. Cold stress at meiosis could become more common in future years because climate change is expected to result in mild winters and warm springs, which are likely to speed up plant development prematurely, resulting in exposure of vulnerable plant tissues and organs to subsequent late season frosts. This occurred in 2007 when severe low temperatures following a period of above average temperatures caused widespread damage to agriculture in the USA (Gu et al. 2008). Models also predict an increase in frequency of heat stress at meiosis (Semenov et al. 2014). Our experiments have shown that even a relatively short period of high temperature at a critical stage of wheat development is sufficient for meiosis to be significantly affected. If current weather trends continue, it will become increasingly important to cultivate both heat-tolerant varieties and cold-tolerant varieties.
Heat tolerance is thought to be a complex trait in plants and is likely to be under the control of multiple genes (Barnabás et al. 2008), but we still know relatively little about the role of individual genes controlling temperature tolerance in wheat (Mullarkey and Jones 2000). Identification of TaDmc1 as a candidate gene that can stabilise chromosome synapsis against extremes of temperature may allow wheat breeders to exploit this information and can provide markers that will allow them to identify and select hexaploid wheat genotypes that carry low (and high) temperature tolerance alleles at this locus.
References
Alabdullah AK, Borrill P, Martin AC, Ramirez-Gonzalez RH, Hassani-Pak K, Uauy C, Shaw P, Moore G (2019) A co-expression network in hexaploid wheat reveals mostly balanced expression and lack of significant gene loss of homeologous meiotic genes upon polyploidization. Front Plant Sci 10:1325. https://doi.org/10.3389/fpls.2019.01325
Armstrong SJ, Caryl AP, Jones GH, Franklin FCH (2002) Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci 115:3645–3655. https://doi.org/10.1242/jcs.00048
Asseng S, Ewert F, Martre P, Rotter RP, Lobell DB, Cammarano D, Kimball BA, Ottman MJ, Wall GW, White JW, Reynolds MP, Alderman PD, Prasad PVV, Aggarwal PK, Anothai J, Basso B, Biernath C, Challinor AJ, De Sanctis G, Doltra J, Fereres E, Garcia-Vila M, Gayler S, Hoogenboom G, Hunt LA, Izaurralde RC, Jabloun M, Jones CD, Kersebaum KC, Koehler AK, Muller C, Naresh Kumar S, Nendel C, O’Leary G, Olesen JE, Palosuo T, Priesack E, Eyshi Rezaei E, Ruane AC, Semenov MA, Shcherbak I, Stockle C, Stratonovitch P, Streck T, Supit I, Tao F, Thorburn PJ, Waha K, Wang E, Wallach D, Wolf J, Zhao Z, Zhu Y (2015) Rising temperatures reduce global wheat production. Nat Clim Change 5:143–147. https://doi.org/10.1038/nclimate2470
Bannister LA, Pezza RJ, Donaldson JR, de Rooij DG, Schimenti KJ, Camerini-Otero RD, Schimenti JC (2007) A dominant, recombination-defective allele of Dmc1 causing male-specific sterility. PLoS Biol 5:e105. https://doi.org/10.1371/journal.pbio.0050105
Barber HM, Carney J, Alghabari F, Gooding MJ (2015) Decimal growth stages for precision wheat production in changing environments? Ann Appl Biol 166:355–371
Barnabás B, Jäger K, Fehér A (2008) The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ 31:11–38. https://doi.org/10.1111/j.1365-3040.2007.01727.x
Bayliss MW, Riley R (1972a) An analysis of temperature-dependent asynapsis in Triticum aestivum. Genet Res 20:193–200. https://doi.org/10.1017/S0016672300013707
Bayliss MW, Riley R (1972b) Evidence of premeiotic control of chromosome pairing in Triticum aestivum. Genet Res 20:201–212
Bennett MD, Chapman V, Riley R (1971) The duration of meiosis in pollen mother cells of wheat, rye and Triticale. Proc R Soc Lond B 178:259–275
Bennett MD, Smith JB, Kemble R (1972) The effect of temperature on meiosis and pollen development in wheat and rye. Can J Genet Cytol 14:615–624
Bennett MD, Rao MK, Smith JB, Bayliss MW (1973) Cell development in the anther, the ovule, and the young seed of Triticum aestivum L. var. Chinese Spring. Philos Trans R Soc B 266:39–81
Bilgir C, Dombecki CR, Chen PF, Villeneuve AM, Nabeshima K (2013) Assembly of the synaptonemal complex is a highly temperature-sensitive process that is supported by PGL-1 during Caenorhabditis elegans meiosis. G3 (Bethesda) 3:585–595. https://doi.org/10.1534/g3.112.005165
Bishop DK, Park D, Xu L, Kleckner N (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69:439–456. https://doi.org/10.1016/0092-8674(92)90446-J
Boden SA, Langridge P, Spangenberg G, Able JA (2009) TaASY1 promotes homologous chromosome interactions and is affected by deletion of Ph1. Plant J 57:487–497
Bolser D, Staines DM, Pritchard E, Kersey P (2016) Ensembl plants: integrating tools for visualizing, mining, and analyzing plant genomics data. In: Edwards D (ed) Plant bioinformatics: methods in molecular biology, vol 1374. Humana Press, New York
Bomblies K, Higgins JD, Yant L (2015) Meiosis evolves: adaptation to external and internal environments. New Phytol 208:306–323. https://doi.org/10.1111/nph.13499
Chapman V, Miller TE (1981) The location of a gene affecting meiotic chromosome pairing at low temperature in Triticum aestivum. Z Pflanzenzüchtg 86:50–55
Colas I, Macaulay M, Higgins JD, Phillips D, Barakate A, Posch M, Armstrong SJ, Franklin FCH, Halpin C, Waugh R, Ramsay L (2016) A spontaneous mutation in MutL-homolog 3 (HvMLH3) affects synapsis and crossover resolution in the barley desynaptic mutant des10. New Phytol 212:693–707. https://doi.org/10.1111/nph.14061
Colas I, Barakate A, Macaulay M, Schreiber M, Stephens J, Vivera S, Halpin C, Waugh Ramsay L (2019) desynaptic5 carries a spontaneous semi-dominant mutation affecting Disrupted Meiotic cDNA 1 in barley. J Exp Bot 70:2683–2698. https://doi.org/10.1093/jxb/erz080
Collins M, Knutti R, Arblaster J, Dufresne J-L, Fichefet T, Friedlingstein P, Gao X, Gutowski WJ, Johns T, Krinner G, Shongwe M, Tebaldi C, Weaver AJ, Wehner M (2013) Long-term climate change: projections, commitments and irreversibility. In: Stocker TF, Qin D, Plattner G-K, Tignor M, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, Midgley PM (eds) Climate change 2013: the physical science basis. Contribution of working group I to the fifth assessment report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge
Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux M-P (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11:1623–1634. https://doi.org/10.1105/tpc.11.9.1623
De Storme N, Geelen D (2014) The impact of environmental stress on male reproductive development in plants: biological processes and molecular mechanisms. Plant Cell Environ 37:1–18
Devisetty UK (2010) Molecular investigation of RAD51 and DMC1 homoeologous genes of hexaploid wheat (Triticum aestivum L.). PhD thesis, University of Nottingham. http://eprints.nottingham.ac.uk/13340/1/523039.pdf. Accessed 17 Dec 2019
Devisetty UK, Mayes K, Mayes S (2010) The RAD51 and DMC1 homoeologous genes of bread wheat: cloning, molecular characterization and expression analysis. BMC Res Notes 3:245
Ding Z, Wang T, Chong K, Bai S (2001) Isolation and characterization of OsDMC1, the rice homologue of the yeast DMC1 gene essential for meiosis. Sex Plant Reprod 13:285. https://doi.org/10.1007/s004970100065
Dowrick GJ (1957) The influence of temperature on meiosis. Hered 11:37–49. https://doi.org/10.1038/hdy.1957.4
Draeger T, Moore G (2017) Short periods of high temperature during meiosis prevent normal meiotic progression and reduce grain number in hexaploid wheat (Triticum aestivum L.). Theor Appl Genet 130:1785–1800. https://doi.org/10.1007/s00122-017-2925-1
Elliott CG (1955) The effect of temperature on chiasma frequency. Hered 9:385–398
Endo TR, Gill BS (1996) The deletion stocks of common wheat. J Hered 87:295–307. https://doi.org/10.1093/oxfordjournals.jhered.a023003
Fischer RA (1985) Number of kernels in wheat crops and the influence of solar radiation and temperature. J Agric Sci 105:447–461. https://doi.org/10.1017/S0021859600056495
Fischer RA, Maurer R (1976) Crop temperature modification and yield potential in a dwarf spring wheat. Crop Sci 16:855–859
Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S, Colas I, Moore G (2006) Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439:749–752. https://doi.org/10.1038/nature04434
Gu L, Hanson PJ, Post WM, Kaiser DP, Yang B, Nemani R, Pallardy SG, Meyers T (2008) The 2007 eastern US spring freeze: increased cold damage in a warming world? Bioscience 58:253–262. https://doi.org/10.1641/B580311
Hayter AM (1969) Cytogenetics and cytochemistry of wheat species. PhD thesis, Cambridge University, Cambridge
Hayter AM, Riley R (1967) Duplicate genetic activities affecting meiotic chromosome pairing at low temperatures in Triticum. Nature 216:1028–1029. https://doi.org/10.1038/2161028a0
Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FCH (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev 19:2488–2500
Higgins JD, Perry RM, Barakate A, Ramsay L, Waugh R, Halpin C, Armstrong SJ, Franklin FCH (2012) Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell 24:4096–4109. https://doi.org/10.1105/tpc.112.102483
International Wheat Genome Sequencing Consortium (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. https://doi.org/10.1126/science.aar7191
Ji H, Xiao L, Xia Y, Song H, Liu B, Cao W, Zhu Y, Liu L (2017) Effects of jointing and booting low temperature stresses on grain yield and yield components in wheat. Agric For Meteorol 243:33–42
Khoo KHP, Able AJ, Able JA (2012) The isolation and characterisation of the wheat molecular ZIPper I homologue, Ta ZYP1. BMC Res Notes 5:106. https://doi.org/10.1186/1756-0500-5-106
Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, Kersey P, Flicek P (2011) Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database: 2011, bar030. https://doi.org/10.1093/database/bar030
Lambing C, Franklin FCH, Wang C-JR (2017) Understanding and manipulating meiotic recombination in plants. Plant Physiol 173:1530–1542. https://doi.org/10.1104/pp.16.01530
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Law CN (1999) Sterility in winter wheat: review of occurrence in different varieties and possible causes. HGCA Res Rev 44:1–108
Martín AC, Shaw P, Phillips D, Reader S, Moore G (2014) Licensing MLH1 sites for crossover during meiosis. Nat Commun 5:1–5
Martín AC, Rey M-D, Shaw P, Moore G (2017) Dual effect of the wheat Ph1 locus on chromosome synapsis and crossover. Chromosoma 126:669–680. https://doi.org/10.1007/s00412-017-0630-0
Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 15:17–29. https://doi.org/10.1093/hmg/ddl046
Mullarkey M, Jones P (2000) Isolation and analysis of thermotolerant mutants of wheat. J Exp Bot 51:139–146. https://doi.org/10.1093/jexbot/51.342.139
Neale MJ, Keeney S (2006) Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442:153–158
Page SL, Hawley RS (2004) The genetics and molecular biology of the synaptonemal complex. Ann Rev Cell Dev Biol 20:525–558
Pallotta MA, Warner P, Fox RL, Kuchel H, Jefferies SJ, Langridge P (2003) Marker assisted wheat breeding in the southern region of Australia. Proc 10th Int Wheat Genet Symp 2:789–791
Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA, Schimenti JC (1998) Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell 1:697–705
Porter JR, Gawith M (1999) Temperatures and the growth and development of wheat: a review. Eur J Agron 10:23–36. https://doi.org/10.1016/S1161-0301(98)00047-1
Qin D, Wu H, Peng H, Yao Y, Ni Z, Li Z, Zhou C, Sun Q (2008) Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using Wheat Genome Array. BMC Genom 9:432. https://doi.org/10.1186/1471-2164-9-432
Queiroz A, Mello-Sampayo T, Viegas WS (1991) Identification of low temperature stabilizing genes, controlling chromosome synapsis or recombination, in short arms of chromosomes from the homoeologous group 5 of Triticum aestivum. Hereditas 115:37–41. https://doi.org/10.1111/j.1601-5223.1991.tb00344.x
Ramírez-González RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, Davey M, Jacobs J, van Ex F, Pasha A, Khedikar Y, Robinson SJ, Cory AT, Florio T, Concia L, Juery C, Schoonbeek H, Steuernagel B, Xiang D, Ridout CJ, Chalhoub B, Mayer KFX, Benhamed M, Latrasse D, Bendahmane A, Wulff BBH, Appels R, Tiwari V, Datla R, Choulet F, Pozniak CJ, Provart NJ, Sharpe AG, Paux E, Spannagl M, Bräutigam A, Uauy C, International Wheat Genome Sequencing Consortium (2018) The transcriptional landscape of polyploid wheat. Science 361:662. https://doi.org/10.1126/science.aar6089
Rearick D, Prakash A, McSweeny A, Shepard SS, Fedorova L, Fedorov A (2011) Critical association of ncRNA with introns. Nucleic Acids Res 39:2357–2366. https://doi.org/10.1093/nar/gkq1080
Rey MD, Martín AC, Higgins J, Swarbreck D, Uauy C, Shaw P, Moore G (2017) Exploiting the ZIP4 homologue within the wheat Ph1 locus has identified two lines exhibiting homoeologous crossover in wheat-wild relative hybrids. Mol Breed 37:95. https://doi.org/10.1007/s11032-017-0700-2
Rey MD, Martin AC, Smedley M, Hayta S, Harwood W, Shaw P, Moore G (2018) Magnesium increases homoeologous crossover frequency during meiosis in Zip4 (Ph1 gene) mutant wheat-wild relative hybrids. Front Plant Sci 9:509. https://doi.org/10.3389/fpls.2018.00509
Riley R (1966) Genotype-environmental interaction affecting chiasma frequency in Triticum aestivum. In: Darlington CD, Lewis KR (eds) Chromosomes today, vol 1. Oliver & Boyd, Edinburgh, pp 57–64
Riley R, Chapman V, Young RM, Bellfield AM (1966) Control of meiotic chromosome pairing by the chromosomes of homoeologous group 5 of Triticum aestivum. Nature 212:1475–1477
Roberts MA, Reader SM, Dalgliesh C, Miller TE, Foote TN, Fish LJ, Snape JW, Moore G (1999) Induction and characterization of Ph1 wheat mutants. Genetics 153:1909–1918
Saini HS, Aspinall D (1982) Abnormal sporogenesis in wheat (Triticum aestivum L.) induced by short periods of high temperature. Ann Bot 49:835–846
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675
Sears ER (1977) An induced mutant with homoeologous pairing in common wheat. Can J Genet Cytol 19:585–593
Semenov MA, Stratonovitch P, Alghabari F, Gooding MJ (2014) Adapting wheat in Europe for climate change. J Cereal Sci 59:245–256. https://doi.org/10.1016/j.jcs.2014.01.006
Somers DJ, Isaac P, Edwards K (2004) A high-density wheat microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114. https://doi.org/10.1007/s00122-004-1740-7
Steinmeyer FT, Lukac M, Reynolds MP, Jones HE (2013) Quantifying the relationship between temperature regulation in the ear and floret development stage in wheat (Triticum aestivum L.) under heat and drought stress. Funct Plant Biol 40:700–707. https://doi.org/10.1071/FP12362
Szurman-Zubrzycka M, Brygida B, Stolarek-Januszkiewicz M, Kwaśniewska J, Szarejko I, Gruszka D (2019) The dmc1 mutant allows an insight into the DNA double-strand break repair during meiosis in barley (Hordeum vulgare L.). Front Plant Sci 10:761. https://doi.org/10.3389/fpls.2019.00761
Thakur P, Kumar S, Malik JA, Berger JD, Nayyar H (2010) Cold stress effects on reproductive development in grain crops: an overview. Environ Exp Bot 67:429–443. https://doi.org/10.1016/j.envexpbot.2009.09.004
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tottman DR (1987) The decimal code for the growth stages of cereals, with illustrations. Ann Appl Biol 110:441–454. https://doi.org/10.1111/j.1744-7348.1987.tb03275.x
Trnka M, Rötter RP, Ruiz-Ramos M, Kersebaum KC, Olesen JE, Žalud Z, Semenov MA (2014) Adverse weather conditions for European wheat production will become more frequent with climate change. Nat Clim Change 4:637–643. https://doi.org/10.1038/NCLIMATE2242
Wang H, Hu Q, Tang D, Liu X, Du G, Shen Y, Li Y, Cheng Z (2016) OsDMC1 is not required for homologous pairing in rice meiosis. Plant Physiol 171:230–241. https://doi.org/10.1104/pp.16.00167
Wardlaw IF, Dawson IA, Munibi P, Fewster R (1989) The tolerance of wheat to high temperatures during reproductive growth. I. Survey procedures and general response patterns. Aust J Agric Res 40:1–13. https://doi.org/10.1071/AR9890001
Winfield MO, Allen AM, Burridge AJ, Barker GLA, Benbow HR, Wilkinson PA, Coghill J, Waterfall C, Davassi A, Scopes G, Pirani A, Webster T, Brew F, Bloor C, King J, West C, Griffiths S, King I, Bentley AR, Edwards KJ (2016) High density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol J 14:1195–1206. https://doi.org/10.1111/pbi.12485
Yazawa T, Nakayama Y, Fujimoto K, Matsuda Y, Abe K, Kitano T, Abe S-I, Yamamoto T (2003) Abnormal spermatogenesis at low temperatures in the Japanese red-bellied newt, Cynops pyrrhogaster: possible biological significance of the cessation of Spermatocytogenesis. Mol Reprod Dev 66:60–66. https://doi.org/10.1002/mrd.10328
Yoshida K, Kondoh G, Matsuda Y, Habu T, Nishimune Y, Morita T (1998) The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell 1:707–718
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421
Acknowledgements
We are grateful to TR Endo for providing seed of the Chinese Spring 5DL terminal deletion lines; The International Atomic Energy Agency, Vienna, for gamma irradiating the Chinese Spring seed; Philippa Borrill for the functional annotation data; Luzie Wingen for the Python Data matcher script for the alignment of data sets.
Funding
This work was supported by the UK Biological and Biotechnology Research Council (BBSRC) through a grant as part of the ‘Designing Future Wheat’ (DFW) Institute Strategic Programme (BB/P016855/1) and response mode Grant (BB/R0077233/1). MD-R thanks the contract ‘Ayudas Juan de la Cierva-Formación (FJCI-2016-28296)’ of the Spanish Ministry of Science, Innovation and Universities.
Author information
Authors and Affiliations
Contributions
TD generated the gamma deletion lines, phenotyped the Chinese Spring lines at high and low temperature and identified the ttmei1 deletion mutant. TD and AP carried out the KASP genotyping. MDR scored chromosome crossover and performed the statistical analysis. AM carried out the immunolocalisation experiments and produced the immunolocalisation figure. AKA identified putative meiosis-specific genes in the 4-Mb deleted region of ttmei1, carried out the sequence alignments and produced the gene expression figures; TD produced all other figures and wrote the manuscript. PS provided a critique of the approach and contributed to discussion of the results. GM provided the concept, provided thoughts and guidance and revised and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Steven S. Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Suppl. Fig. 1
Gene expression patterns of the three hexaploid wheat TaDmc1 homeologs, TraesCS5A02G133000 (TaDmc1-A1) on chromosome 5A, TraesCS5B02G131900 (TaDmc1-B1) on 5B and TraesCS5D02G141200 (TaDmc1-D1) on 5D in different tissue types, based on the 876 RNASeq samples available in the wheat expression browser (http://www.wheat-expression.com). TaDmc1 expression levels are higher in meiotically active tissues than non-meiotic tissues. In meiotic anthers, expression levels of TaDmc1-D1 are higher than those of TaDmc1-A1 and TaDmc1-B1 (PDF 484 kb)
Suppl. Fig. 2
Multiple alignment of the coding sequences (CDs) of the three hexaploid wheat Dmc1 homeologs: TraesCS5A02G133000 (TaDmc1-A1) on chromosome 5A, TraesCS5B02G131900 (TaDmc1-B1) on 5B and TraesCS5D02G141200 (TaDmc1-D1) on 5D. SNPs are shown in red text on a grey background (PDF 103 kb)
Suppl. Fig. 3
Multiple alignment of the DNA sequences of the three hexaploid wheat Dmc1 homeologs: TraesCS5A02G133000 (TaDmc1-A1) on chromosome 5A, TraesCS5B02G131900 (TaDmc1-B1) on 5B and TraesCS5D02G141200 (TaDmc1-D1) on 5D (DOCX 21 kb)
Suppl. Fig. 4
Multiple sequence alignment of the promoter regions of the three hexaploid wheat Dmc1 homeologs: TraesCS5A02G133000 (TaDmc1-A1) on chromosome 5A, TraesCS5B02G131900 (TaDmc1-B1) on 5B and TraesCS5D02G141200 (TaDmc1-D1) on 5D. The promoter regions include the 1500 nucleotides downstream of the start codon of each of the TaDmc1 homeologs. Note the large insertion mutation (163 nt in size) at position 430 downstream of the start codon of TaDmc1-B1 (PDF 165 kb)
Suppl. Fig. 5
Multiple alignment of Dmc1 DNA sequences from T. aestivum and its diploid and tetraploid ancestors, T. urartu (AA), Ae. tauschii (DD) and T. dicoccoides (AABB); note the large insertion of 294 bp in the B-genome copy of the T. aestivum gene (DOCX 33 kb)
Suppl. Fig. 6
DMC1 transcripts in T. aestivum and its diploid and tetraploid ancestors (as predicted in Ensembl Plants) (DOCX 982 kb)
Suppl. Fig. 7
Multiple alignment of DMC1 amino-acid sequences from T. aestivum (Ta) and its diploid and tetraploid ancestors: the A-genome donor, T. urartu (Tu_A), the D-genome donor, Ae. tauschii (Aet_D) and the AABB genome progenitor, T. dicoccoides (Td_A and Td_B). Ae tauschii and T. dicoccoides have multiple transcripts of their Dmc1 genes due to alternative splicing variants, but this alignment only includes transcripts that are most similar to those of T. aestivum. Numbers after the dots refer to the transcript number. Large red triangle indicates the single amino-acid substitution in TaDMC1-D1 that may confer low temperature tolerance in wheat. Note the conservation of this substitution in the ancestral D-genome of Ae. tauschii. Small blue triangles show the positions of three other amino-acid substitutions in TaDMC1-B1, and a small green triangle shows the position of a substitution in TaDMC1-A1 that is also conserved in the ancestral copies (RTF 101 kb)
Suppl. Table 1
The effects of three different temperature treatments on meiotic metaphase I chromosomes of individual Chinese Spring (CS) wild-type and 22-F5 (ttmei1) mutant plants. The mean numbers of univalents, ring and rod bivalents, trivalents, tetravalents and pentavalents were scored along with chiasma frequency scored as single or double crossovers. Standard deviation (SD) and standard error (SE) are shown, as well as the maximum (max) and minimum (min) number of chromosomes with a particular conformation (PDF 675 kb)
Suppl. Table 2
Expression values (TPM) of all genes inside the Dmc1 deletion region in 876 samples from different tissue types (XLSX 246 kb)
Suppl. Table 3
Dmc1 sequence identity matrices. a) Percentages of nucleotide sequence identity for the three TaDmc1 homeologs: TraesCS5A02G133000 (TaDmc1-A1) on chromosome 5A, TraesCS5B02G131900 (TaDmc1-B1) on 5B and TraesCS5D02G141200 (TaDmc1-D1) on 5D; b) percentages of amino-acid sequence identity for DMC1 proteins in wheat (TaDMC1-A1, TaDMC1-B1 and TaDMC1-D1), barley (HvDMC1) and rice (OsDMC1A and OsDMC1B); c) percentages of nucleotide sequence identity for Dmc1 homeologs in T. aestivum (TaDmc1) and its progenitor genomes: T. urartu (TuDmc1-A), Ae. tauschii (AetDmc1-D) and T. dicoccoides A genome (TdDmc1-A) and B genome (TdDmc1-B); d) percentages of amino-acid sequence identity for DMC1 proteins in T. aestivum (TaDMC1) and its progenitor genomes: T. urartu (TuDMC1-A), Ae. tauschii (AetDMC1-D) and T. dicoccoides A genome (TdDMC1-A) and B genome (TdDMC1-B). Two different splice variants each are shown for TdDMC1-A, TdDMC1-B and AetDMC1-D. Note that of all the amino-acid sequences of T. aestivum and its progenitors, T. urartu is the most divergent (XLSX 14 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Draeger, T., C. Martin, A., Alabdullah, A.K. et al. Dmc1 is a candidate for temperature tolerance during wheat meiosis. Theor Appl Genet 133, 809–828 (2020). https://doi.org/10.1007/s00122-019-03508-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03508-9