Abstract
Key message
We report growth habit profiling following SEM, genetic mapping and QTL analysis. Highlighted CcTFL1 , a candidate for determinacy in pigeonpea, since an Indel marker derived from this gene co-segregated with Dt1 locus.
Abstract
Pigeonpea (Cajanus cajan) is one of the most important legume crops grown in arid and semi-arid regions of the world. It is characterized with few unique features compared with other legume species, such as Lotus, Medicago, and Glycine. One of them is growth habit, an important agronomic trait. In the present study, identification of mutations affecting growth habit accompanied by a precise analysis of phenotype has been done which will shed more light upon developmental regulation in pigeonpea. A genetic study was conducted to examine the inheritance of growth habit and a genotyping by sequencing (GBS)-based genetic map constructed using F2 mapping population derived from crossing parents ICP 5529 and ICP 11605. Inheritance studies clearly demonstrated the dominance of indeterminate (IDT) growth habit over determinate (DT) growth habit in F2 and F2:3 progenies. A total of 787 SNP markers were mapped in the genetic map of 1454 cM map length. Growth habit locus (Dt1) was mapped on the CcLG03 contributing more than 61% of total phenotypic variations. Subsequently, QTL analysis highlighted one gene, CcTFL1, as a candidate for determinacy in pigeonpea, since an Indel marker derived from this gene co-segregated with the Dt1 locus. Ability of this Indel-derived marker to differentiate DT/IDT lines was also validated on 262 pigeonpea lines. This study clearly demonstrated that CcTFL1 is a candidate gene for growth habit in pigeonpea and a user-friendly marker was developed in the present study which will allow low-cost genotyping without need of automation.
Similar content being viewed by others
Introduction
The growth habit can differ considerably among cultivars for a given crop. Some cultivars have a vegetative terminal bud, which allows the genotype to grow in height and spread under adequate conditions. Such habit is known as an ‘indeterminate’ (IDT) type. The other form of growth habit is designated as ‘determinate’ (DT) type. The determinate genotypes are relatively short in stature and possess reproductive terminal buds. Such genotypes when reach flowering stop growing in height. Pigeonpea is an important pulse crop which has gained economic importance in developing world, especially in Asia and Africa due to its continuous increase in demand. It is suggested that pigeonpea was domesticated from its wild species relative, Cajanus cajanifolius approximately >3500 years ago (Royes 1976). In wild relative species of pigeonpea, the growth habit is generally indeterminate and the genotype flowers under short-day conditions. Whereas, variability for growth habit exists in the case of cultivated pigeonpea, i.e., some genotypes are IDT type and some genotypes are DT type. On the other hand, some genotypes are also intermediate between the IDT and DT types. These genotypes are called semi-determinate (SDT) types that ultimately contribute directly to traits such as grain yield, harvesting, and seed production. Both the major types of growth habits have their own advantages and limitations in pigeonpea. For instance, DT type genotypes mature rapidly and have a shortened flowering period, allowing earlier maturity and ease of mechanical harvesting. Whereas, control of damage in the case of insect attacks, higher seed rate, etc. is among the limiting factors in popularization of DT genotypes. The DT growth habit in pigeonpea was reported controlled by a single recessive gene (Kapoor and Gupta 1991). Alternative growth habit, i.e., IDT, is a dominant trait and has many advantages which are preferred by pigeonpea growers such as multiple branching, higher yields, easy insect control, lower seed rate, low inputs, etc. However, tall habit of IDT phenotypes and continuous flowering further hampered the mechanization efforts and synchronized harvesting.
The growth habit has been studied in a number of plant species to understand the genetic and genomic controls. Identification of genetic loci underlying growth habit will allow a better understanding of the domestication process and will enable faster, manipulation of growth habit, and flowering time in future breeding efforts. Like many traits, detailed characterization for growth habit at genomic level was initiated in Arabidopsis, and later, orthologous genes were studied in other crops species such as soybean (Tian et al. 2010), pea (Foucher et al. 2003), common bean (Kwak et al. 2008), pigeonpea (Mir et al. 2014), etc. In Arabidopsis, two opposing pathways exist including genes APETALA1 (AP1) which activate flowering initiation and TERMINAL FLOWER1 (TFL1) as a repressor for floral initiation. Previously candidacy of homologous genes from the above-mentioned pathways was tested in other crop species following genetic linkage analysis, candidate gene association analysis, heterologous transformation, etc. (Kwak et al. 2008; Liu et al. 2010; Repinski et al. 2012; Tian et al. 2010; Foucher et al. 2003). In the case of pigeonpea, few markers associated with determinacy were identified (Mir et al. 2012), and following candidate gene analysis CcTFL1, a homolog of Arabidopsis gene TFL1 was reported as likely candidate gene for growth habit (Mir et al. 2014).
To characterize growth habit in pigeonpea, we have followed a systematic approach which includes microscopic characterization of mutations affecting inflorescence ontogeny, understanding the inheritance of growth habit, genotyping by sequencing (GBS)-based genetic mapping, and validation of identified associated genomic segments in diverse genetic stocks. This study also presents validation to support our hypothesis that CcTFL1 is the gene underlying growth habit in pigeonpea. In addition, a user-friendly Indel marker has been developed from CcTFL1 for quick and accurate manipulation of growth habit in pigeonpea breeding program.
Materials and methods
Morphological characterization and scanning electron microscopy
The following genotypes were studied for morphological descriptions: ICPL 20338, ICPL 87091 (determinate), ICPL 20325, and ICPL 88039 (indeterminate). Plants were grown in glasshouse conditions. In addition, line ICP 7035 and some related genera were observed in the pigeonpea wild relatives’ germplasm collection. Plant material (apices and floral buds) was fixed and stored in 70% ethanol. For further scanning electronic microscope (SEM) analysis, the specimens were dissected under a stereomicroscope, critical-point dried, mounted on metal stubs and coated with Au + Pd in a sputter coater Eiko IB-3. Specimens were visualized using an SEM CamScan-S2 (Cambridge Instruments, UK; secondary electron image regime) with accelerating voltage of 20 kV.
Field experiment and phenotyping
One F2 mapping population consisting of 202 individuals segregating for growth habit (DT/IDT) was developed by crossing parental lines, namely, ICP 5529 (IDT) and ICP 11605 (DT). The sowing was done in 4-m-long rows with row-to-row and plant-to-plant spacing of 75 and 30 cm, respectively. The F3 seeds derived from the F2 plants were also sown in the field in progeny test rows. F3 seeds from F2 plants were sown in a single 4-m-long row. Plants were phenotyped on single plant basis for indeterminate or determinate growth habit after the onset of flowering as described in Mir et al. (2014).
DNA isolation and genotyping by sequencing (GBS)
Total genomic DNA was isolated from 188 F2s and parents’ seedlings 2 weeks after germination using NucleoSpin plant extraction kit (MACHEREY–NAGEL, USA). DNA was also isolated from a set of 262 genotypes for marker validation. The quality and quantity of DNA were checked on 0.8% agarose gel. GBS approach was used for single nucleotide polymorphism (SNP) discovery and genotyping the F2 population. In summary, 10 ng of total genomic DNA from each sample was digested using ApeKI endonuclease. The digested DNA was ligated with barcoded adaptors using T4 DNA ligase. Ligated products from each sample were mixed in equal proportion to construct the GBS libraries. Furthermore, GBS libraries were amplified, purified, and used for sequencing on HiSeq 2500 platform (Illumina Inc, San Diego, CA, USA) as mentioned in Jaganathan et al. (2015).
SNP genotyping
The sequence reads generated on HiSeq 2500 were used for SNP identification and genotyping using GBS analysis pipeline implemented in TASSEL v4.0. The barcode containing reads were sorted, de-multiplexed, and trimmed to first 64 bases starting from enzyme cut site. Reads containing ‘N’ within first 64 bases were discarded. The good quality reads (called as tags) were aligned against the draft genome sequence of pigeonpea (Varshney et al. 2012) using Burrows–Wheeler Alignment tool (BWA) (Li and Durbin 2009). The alignment file was processed through GBS analysis pipeline for SNP calling and genotyping. SNPs with contrasting alleles in parental genotypes and having <30% missing data were retained for study. Furthermore, imputation of missing data was carried out using FSFHap algorithm implemented in TASSEL v4.0. The imputed SNPs were subjected to minor allele frequency (MAF) cutoff of 0.2 to remove missing data and such filtered SNPs were used for genetic mapping and QTL analysis.
Primer design and polymerase chain reaction (PCR)
Information on CcTFL1 gene was obtained from Mir et al. (2014). To find sequence variations, CcFTL1 gene sequence was aligned onto crossing parental of mapping population, i.e., ICP 5529 (IDT) and ICP 11605 (DT) using Asha reference genome sequence. A 10 bp deletion was identified on CcLG03 at position 20698771 bp in DT genotype as compared to IDT genotype. One primer pair was designed using the online primer design software Primer3 plus. The primer pair used for amplification include: CcLG03_ 20698771_F (CAT GGC CAT TGT AGA CTT GCT (21 bp) and CcLG03_ 20698771_R (TCA CAG CAG GAT CAT CGA GT (20 bp).
PCR reaction was conducted in a total volume of 30 μL containing 21.9 μL of ddH20, 2.0 μL of 10 × KAPA Taq polymerase buffer (containing 25 mM MgCl2), 2.0 μL of 10 mM dNTPs, 10 pmol/μL of each of the forward and reverse primers, 0.06 μL of KAPA Taq polymerase, and 2.0 μL of 20 ng/μL gDNA. A touch-down PCR was used as follows: initial denaturation at 95 °C for 5 min followed by two annealing cycles: (1) 5 cycles consisting of 94 °C for 15 s, 62 °C for 20 s, and 72 °C for 30 s and (2) 35 cycles consisting of 94 °C for 15 s, 54 °C for 30 s, and 72 °C for 30 s and a final extension at 72 °C for 20 min and hold at 4 °C for infinity. PCR products were run in 3.5% Nusieve agarose gel. Gels were stained with ethidium bromide and visualized under UV light in a transilluminator.
Data analysis
The F2 and F3 phenotypic segregation data were subjected to Chi-square goodness-of-fit analysis to test conformation to expected Mendelian segregation ratios. Single marker regression analysis was carried out using F2 phenotypes as dependent variables and the F2 marker genotypes as independent variables in Microsoft Excel 2013. The CcTFL1 gene marker data were incorporated into a GBS-derived SNP-based genetic map using JoinMap 4.1 with the following settings: independence LOD 2.5 to 10.0, regression mapping algorithm, recombination frequency ≥0.49 and a χ 2 jump threshold for removal of loci set 5.0, and a ‘‘Ripple’’ after adding one marker into the map. The inter-marker distances calculated from the JoinMap program were used to construct a genetic map which was displayed using MAPCHART version 2.2. Inclusive composite interval mapping (ICIM) was conducted using the version 4.0 of QTL IciMapping software.
Results
Scanning electron microscopy-based characterization of IDT and DT genotypes
Stem architecture
IDT genotypes have been characterized with cryptocotylar hypogeal germination, i.e., cotyledons remain in a seed coat not emerging from ground. The first aerial node bears two opposed single-leaflet leaves. The second node bears two opposed leaves of either the same morphology as the first, or one of them was of single leaflet while another trifoliate, as on higher nodes. In few plants, a phyllotaxis was alternate, since the second node, but in this case, all leaves except for the first node were trifoliate. After transition to alternating phyllotaxis, stems of pigeonpea exhibit a handedness. Leaves were arranged in spirals, either right (clockwise) or left (counterclockwise) (Figs. 1, 2a). The angle between leaves is 137.5°. Although limited samples of each genotype were tested, ratio between right- and left-handed plants seems close to 1:1 (7:6 in ICPL 20325, χ 2 = 0.077, p < 0.05). After producing few leaves with vegetative buds in their axils, plants begin flowering. The first node which produces a lateral flower-bearing axis would further be designated as a node of flowering initiation (NFI). An average number of nodes preceding NFI vary in different genotypes (7–10 in ICPL 20325, 8–13 in ICPL 88039). In the case of DT pigeonpea genotypes, peculiarities of germination and vegetative growth were found similar to IDT type. Ratio between right- and left-handed plants was also similar to 1:1 (13:11 in ICPL 20338, χ 2 = 0.167, p < 0.05; 9:11 in ICPL 87091, χ 2 = 0.200, p < 0.05). The number of sterile nodes preceding NFI was variable depending on genotype and a certain plant (12–17 in ICPL 87091, 5–7 in ICPL 20338).
Schematic representations of inflorescence morphology in pigeonpea. A, B—IDT plant: scheme (A) and diagram (B). Arrow (crossed circle on B) continuously proliferating axis, hatched shortened internodes, black arc bract, white arc (black acute arc on B) prophyll, white acute arc frondose leaf, half-grey circle flower (grey half abaxial side), black circle axis with limited growth, circle with dot dormant axillary bud, grey arrows order of flowering, N node of flowering initiation, T terminal inflorescence. C, D—DT plant. E—axillary complex in line ICPL 87091. F—axillary complex in DT plants
Apices of IDT (a, c, d, g) and DT (b, e, f) pigeonpea plants (SEM microphotographs). a Indeterminate vegetative apex (asterisk) of DT plant surrounded with primordia of frondose leaves (L) which comprise right spiral. Arrow order of initiation. b Apex of DT plant with pairs of flowers arising in axils of bracts (b) arranged in left spiral. c Flowers arising on I3 apex (seen between floral primordia). d Pair of flowers on later stage. e Flower pair of DT plant; I3 apex (dot) also produces phyllome (arrow). f Pair of flowers of DT plant with pin-like phyllome protruding between them. g Flower pair (IDT plant), preanthetic stage. Scale bars 50 μm (c–e), 100 μm (a), 200 μm (b), 500 μm (g), 1 mm (f)
Inflorescence structure
IDT pigeonpea genotypes beginning from NFI, every leaf on the main (first order, I1) stem subtends an axillary (second order, or partial) racemose inflorescence (I2, Fig. 1a). A phyllotaxis of floral zone of the main shoot follows the same pattern as a preceding vegetative portion. The I1 stem proliferates unlimitedly. Every axillary partial inflorescence begins with two scale-like persistent prophylls which remain at its base, in an axil of frondose leaf (Fig. 1a). Each prophyll subtends a bud. In C. cajan grown in a glasshouse, these buds usually remain dormant, but one of them was developed more fully than another. This asymmetry indicated that prophylls were arranged in alternating (distichous) way rather than opposed. In C. cajanifolius inflorescences, one or both buds in prophylls’ axils usually unfold producing the third-order racemose inflorescences (I3′, Fig. 1a) similar to one which bears prophylls. A similar phenomenon was observed in C. cajan grown in a field. After producing two prophylls, a partial inflorescence begins initiation of bracts. Bracts were of oblonged ovate shape, with acute tip, densely pubescent. Arrangement (phyllotaxis) of bracts on axis was also spiral, with divergence angle which seems similar to one of the main axis. In many cases, torsion angles on the first- and second-order axes were converse. For example, if the main stem torsion was right, most (usually not all) of the axillary inflorescences would be left. In some plants, this right–left conversion between the first- and second-order axes was perfect, but in most cases, it had exclusions. The third-order racemose inflorescences (I3′), if unfold, often had a torsion converse compared with the second-order inflorescences on which they develop. This regularity was also not strictly followed. On every partial inflorescence, the first bract was initiated closer to the prophyll which subtends more developed bud. This pattern was obeyed rigorously, so chirality of a partial inflorescence can be predicted basing on position of prophylls (Fig. 1b). Every bract subtends a pair of flowers (Fig. 1b) which were initiated at apex as a loaf-shaped meristem primordium (Fig. 2b) which later splits into two floral primordia (Fig. 2b, c). A remaining meristem between them comprises an apex of shortened third-order axis (I3) which stops its proliferation after production of two flowers (Fig. 2d). These flowers seem to develop somewhat asynchronously, i.e., are initiated in successive mode. This phenomenon can also be seen at flowering stage when two flowers in pair open one after another, not simultaneously. Flowers themselves were not subtended by any discernible bracts nor bear bracteoles. Among all examined genotypes, we found only ICPL 87091, in which individual flowers were preceded by small-scaly abscising bracts (Fig. 1e). Heritability of this feature is yet to be studied. Flowers were positioned with their adaxial sides to each other (Fig. 2d, g). The number of nodes on I2 which produce bracts and I3 varies along the shoot together with length of sterile portion of I2. Length of the latter decreases in acropetal direction, as does number of flowers. However, in both field and greenhouse conditions, flowers of distal portions of I2 senesce before opening, so a potential productivity were never achieved and can hardly be estimated precisely.
In DT pigeonpea genotypes as compared with indeterminate genotypes, plants exhibit transition of I1 to production of bracts instead of frondose leaves. These bracts subtend pairs of flowers, like on I3 (Figs. 1c, 2b). A phyllotaxis of terminal inflorescence continues one of the preceding zone of axillary inflorescences (Fig. 1d). In studied DT genotypes, production of additional organs on I3 was observed. Some of I3 apices also produced the third flower or bract-like phyllome (Fig. 2e, f). Such morphogenesis pattern in IDT plants was not detected, so we suppose that these features were associated. The number of flowering nodes in terminal inflorescence always exceeds one in previous axillary pseudoraceme and seems similar to the pseudoraceme which arises in NFI (Fig. 3b). In IDT plants, every successive node produces fewer flowers than preceding one (Fig. 3a).
Inheritance of growth habit
A complete dominance of IDT was observed in F1s generated by crossing parental genotypes, namely, ICP 5529 (IDT) with ICP 11605 (DT). In F2 generation, a total of 202 individuals were phenotyped for IDT and DT growth habits. The F2 population segregated in 3 IDT: 1 DT ratio with probability value of 0.68 suggesting that IDT is controlled by a single dominant gene (Electronic supplementary material Table 1). In F3 generation, pooled analysis of 1597 F3 plants from randomly chosen 126 F2s, 855 F3 plants from 48 F2s segregated for IDT/DT, while 742 F3 from 78 F2s did not segregate. This segregation pattern fit well to the expected ratio of 1 segregating: 1 non-segregating (p = 0.30) (Electronic supplementary material Tables 2 and 3). The variation observed within 48 segregating progenies indicated the presence of a single gene with its dominant allele causing IDT, and segregated in 3 IDT:1 DT ratio (Electronic supplementary material Table 4).
Sequence data and SNPs’ discovery
Parental genotypes (ICP 5529 × ICP 11605) together with 179 from a total of 188 F2s were sequenced, and a total of 305.53 million reads containing 30.85 Gb data were generated. DNA from 9 F2s could not be used for library preparation due to low concentration and quality. From the above-mentioned sequencing data in parental lines, a total of 5.37 million reads containing 0.54 Gb for ICP 5529 and 1.60 million reads containing 0.16 Gb for ICP 11605 were generated. In 179 F2s, the number of reads generated varied from 0.40 to 5.25 millions with an average of 1.66 million per F2 line (Electronic supplementary material Table 5). As a result, generated sequencing data on 179 F2 lines were used for SNPs identification and subsequently parsed to remove heterozygous SNPs in parental genotypes. Following stringent filtering criterion, a total of 12,654 SNPs were identified across 179 F2s.
CcTFL1-derived Indel marker and segregation
To validate our hypothesis of association of CcTFL1 gene to the growth habit in pigeonpea and to develop a user-friendly marker, we took advantage of available CcTFL1 gene sequence from Mir et al. (2014) and re-sequencing data on determinate and indeterminate genotypes (Kumar et al. 2016). As a first instance, CcTFL1 gene sequence was aligned across parental genotypes (ICP 5529 and ICP 11605) of F2 mapping population and reference pigeonpea genome. Sequence alignment has detected five potential variations in CcTFL gene region (Electronic supplementary material Table 6). Out of five sequence variations detected between ICP 5529 and ICP 11605, four were heterozygous in either of the parent. A 10 bp deletion has been identified on CcLG03pseudomolecule in DT genotype ICP 11605 while comparing with IDT genotype ICP 5529. Annotation of identified Indel has shown its active role in creating a frame shift mutation. Further flanking sequences of Indel region were used for primer designing. The primer pair designed for Indel region [CcLG03_ 20698771_F (CAT GGC CAT TGT AGA CTT GCT (21 bp) and CcLG03_ 20698771_R (TCA CAG CAG GAT CAT CGA GT (20 bp)] has been used to genotype F2 population derived from cross ICP 5529 × ICP 11605 (Electronic supplementary material Figure 1). The CcTFL1 fragment containing 10 bp Indel segregated in the F2 population suggesting that CcTFL1 is a candidate gene for growth habit and this marker was termed S3_20698771.
Marker S3_20698771 differentiated all 188 F2 plants into 148 IDT plants and 40 DT plants on agarose gel. All determinate plants were homozygous for the marker S3_20698771. Among indeterminate F2 plants, 107 were heterozygous and 41 were homozygous for the marker S3_20698771. A goodness-of-fit test for a 1 determinate (homozygous):2 indeterminate (heterozygous):1 indeterminate (homozygous) provided a χ 2 value of 3.6 at probability value of 0.16 (Electronic supplementary material Table 7).
SNPs-based genetic map
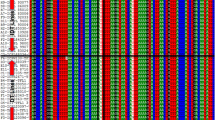
From a total of 12654 SNPs identified, 2935 SNPs showed segregation in 1:2:1 ratio with a cut-off probability value 10−9 and retained for genetic mapping. Further marker genotyping data generated through CcTFL1-derived Indel marker (S3_20698771) on F2 population was also added in SNP data set for constructing genetic map. As segregation distortion from the expected ratio was on higher side in the F2 population, a subgroup of 714 markers with a cut-off probability value ≥0.05 was created and considered as anchor markers for initiating genetic map construction. As a result, a total of 140 markers with probability value ≥0.05 could be mapped in framework genetic map. Furthermore, 647 markers with probability value <0.05 ≥ 10−9 could be integrated into framework genetic map accounting a total of 787 markers in the genetic map with 1454 cM map length (Fig. 4). The highest number of markers was mapped on CcLG11 (249), while the lowest number of markers was mapped to CcLG10 (8). Length of linkage groups varied from 40.5 cM (CcLG10) to 205 cM (CcLG11). Overall, map had 0.54 markers per cM on average (Table 1).
Gentic map and QTLs for the ICP 5529 × CP 11605 mapping population. Bars indicate QTL positions: Red bar is a major QTL with one of the two flanking markers being CcLG03_20698771 deletion marker derived from the CcTFL1 gene (Mir et al. 2014) with the PVE of 61.6, Blue bars represents other major QTLs and Green bar is a minor QTL. All coloured markers are flanking QTL positions
QTLs for growth habit
QTL mapping for growth habit was performed using trait phenotypic data together with marker genotyping data on the F2 mapping population following inclusive composite interval mapping (ICIM). Marker genotyping data included 786 SNPs and one CcTFL1-derived Indel marker. ICIM identified a total of five QTLs for growth habit in pigeonpea. Out of these identified five QTLs, four QTLs were considered as major effect QTLs (explaining more than 10% of phenotypic variation). Whereas one QTL identified on CcLG02 was considered as minor effect QTL explaining 3.9% of phenotypic variation. Among major effect QTLs, two QTLs were located on CcLG03 and one QTL each on CcLG04 and CcLG11. The QTL flanked by S3_24127385 and S3_21274904 markers on CcLG03 explained 12% of phenotypic variation at 4.4 LOD value. Another QTL flanked by S3_20698771 and S3_18430894 markers on CcLG03 explained 61.6% of phenotypic variation at 29.1 LOD value. It is interesting to note that marker S3_20698771 has been derived from an Indel identified in genic region of CcTFL1 and explaining almost two-third of total of phenotypic variation explaining growth habit in pigeonpea. QTL flanked by S4_496463 and S4_487510 markers on CcLG04 explained 13.1% of phenotypic variation at 2.6 LOD value. The fourth major QTL was on CcLG11 flanked by S11_8456082 and S11_44938548 markers explained 14% of phenotypic variation at 4.1 LOD value (Table 2; Fig. 5).
Validation of CcTFL1-derived Indel marker
To validate association of CcTFL1-derived Indel marker (S3_20698771) with growth habit in pigeonpea, a set of 262 genotypes was used. This set comprised of 16 DT, 10 SDT, and 236 IDT genotypes. Primer pairs used to amplify marker S3_20698771, a single amplification product of 200 bp was obtained in all IDT and SDT genotypes. In all the 16 DT genotypes, a single amplification product of 190 bp was obtained using similar primer pairs mentioned above (Electronic supplementary material Figure 2).
Discussion
The apical meristem of shoot is a set of stem cells persistent at the shoot axis. It proliferates and generates leaves with axillary organs, either vegetative shoots or flowers (Bowman et al. 1993). The fate of apical meristem was studied thoroughly in Arabidopsis and depends on generally two sets of genes: first set of genes such as LEAFY (LFY), APETALA1 (AP1) and CAULIFLOWER (CAL) converts meristem into flowers rather than shoots or leaves (Bowman et al. 1993; Gustafson et al. 1994; Kempin et al. 1995; Mandel and Yanofsky 1995). Second set of genes such as TERMINAL FLOWER1 (TFL1) represses flower generation from apical meristem and gives rise to indeterminate growth habit (Bradley et al. 1996). TFL1 gene also contributes to flowering time, influencing phase expansion and inflorescence formation in Arabidopsis (Bradley et al. 1997; Ruiz-García et al. 1997). TFL1 leads down regulation of LFY and AP1 genes, hence prevent their expression in apical meristem. Mutation in TFL1 converts apical meristem to terminal flower and gives rise to determinate type of growth habit. The vegetative and reproductive durations have been significantly shortened in TFL1 mutant plants (Ahn et al. 2006). Due to significance of growth habit trait, a number of attempts have been made to detect the homologous genes of TFL1 and understand its mechanism in different plant species such as brassica, pea, grapevine, lotus, tomato, tobacco, apple, cotton, cucumber, pepper, soybean, common bean, pigeonpea, etc. To characterize growth habit trait in pigeonpea, a comprehensive approach has been followed in the present study.
Pigeonpea is a member of Phaseoleae tribe which is one of the most diverse in legume family. In pigeonpea, bracts on I2 subtend extremely shortened I3 axes which usually bear two flowers, the latter developing in alternate way. Such inflorescence type is called pseudoraceme (Tucker, 1987). Stem apical meristem (SAM) of I3 was seen only during flower initiation (Fig. 2c) and indistinguishable by anthesis. Flowers were initiated in axils of prophylls which arise in transversal position and usually remain cryptic themselves. We found a single genotype in which they develop as bracts; this trait may be heritable. Pigeonpea plants have spiral phyllotaxis. A phyllotaxis torsion of any individual plant may be either right or left. In Arabidopsis, a suite of mutants which cause development of exclusively right- or left-handed plants was isolated (Hashimoto 2002). In pigeonpea, we recorded both types in all examined genotypes, and ratio between plants with different torsion corresponded significantly to 1:1. Although Bahadur and Rao (1981) after analyzing larger sample reported that ratio between different chirality types significantly deviates from 1:1, we propose that this feature has no genetic determination in our studied material. Otherwise, one can hardly imagine any ratio among isogenic plants. A phyllotaxis of terminal inflorescence (I2*, Fig. 1c) continues one of preceding I1 zone. This continuation is evident from both divergence angle and torsion, either right or left. A similar pattern was recorded in DT forms of pea and broad bean (Sinjushin 2013), where the first bract of terminal raceme is usually placed exactly, where the frondose leaf of preceding shoot should develop. A terminal inflorescence (I2*) was larger than preceding lateral ones (I2) (Fig. 3b), as it includes more nodes with axillary I3s. We observed production of additional structures on I3 of DT plants (Fig. 2e, f). These structures may have dorsoventral habit and seemingly comprise phyllomes (bracts). Sometimes, the third flower with subtending bract develops in median plane (Fig. 1f). These structures do not seem truly terminal on I3: the I2 apex is visible (Fig. 2e). In Arabidopsis, gene TFL1 controls both inflorescence determinacy and flowering time, as mutants tfl1 begin flowering significantly earlier than wild-type plants (Hanano and Goto 2011). As it had been shown in pea, gene TFL1 has three orthologs (Foucher et al. 2003). One of them, PsTFL1a, corresponds to gene DETERMINATE (DET) which prevents SAM from production of apical raceme. Another, PsTFL1c, comprises gene LATE FLOWERING (LF) which defines flowering time: NFI in lf mutants has lower number than in LF plants. Thus, two functions present in Arabidopsis become distributed between two TFL1 orthologs in pea.
Dt 1 locus responsible for indeterminacy in pigonpea
Limited information is available to understand the genetic control of growth habit in pigeonpea (Shaw 1936; Reddy and Rao 1974; Saxena and Sharma 1990; Gupta and Kapoor 1991; Gumber and Singh 1997; Sapkal et al. 2015). Through the above-mentioned inheritance studies, it has been proposed that growth habit in pigeonpea is controlled by two dominant genes (Dt 1 and Dt 2 ) with inhibitory interaction. Dt 1 is responsible for IDT phenotype and suppresses the expression of Dt 2 . IDT phenotype can be produced when a plant has either Dt 1_ Dt 2_ or Dt 1_ dt 2 dt 2 combinations. SDT phenotype can be produced when a plant has dt 1 dt 1 Dt 2_ combination. The DT phenotype can only be produced when both the genes are present in recessive and homozygous condition, i.e., dt 1 dt 1 dt 2 dt 2 . In the present study, a segregating population was developed using IDT and DT parental genotypes. The segregation observed (3 IDT: 1 DT) in the present study supports the hypothesis of single dominant gene controlling IDT growth habit in pigeonpea. Similar observations have been made in soybean (Bernard 1972) and broad bean (Brimo 1983). SDT phenotype was also investigated in pigeonpea and appeared recessive with response to IDT (Gupta and Kapoor 1991). Studies conducted in pigeonpea with SDT × IDT cross uncovered dominant epistasis (i.e., 12 IDT:3 SDT:1 DT individuals in F2 progeny) (Gupta and Kapoor 1991; Gumber and Singh 1997). This ratio has also been found in soybean (Bernard 1972), chickpea (Hegde 2011), and tomato (Elkind et al. 1991). In pigeonpea, chickpea, and tomato, SDT has been found recessive trait but dominant in soybean.
Comparisons of soybean with Arabidopsis have shown that Dt1 was an ortholog of TERMINAL FLOWER1 (TFL1) gene (Liu et al. 2010; Tian et al. 2010). TFL1 also involved in the transition from indeterminate phenotype to determinate phenotype (Tian et al. 2010). Therefore, it is worthwhile to map the Dt1 locus in pigeonpea and assess its association with CcTFL1 a homolog of TFL1. It is important to mention that in our previous study, CcTFL1 was identified (Mir et al. 2014); however, its association with Dt1 locus was unknown.
The genetic map developed in the present study covered all the 11 linkage groups of pigeonpea. Total length of this genetic map (1454 cM) was on higher side as compared to previously developed few simple sequence repeats (SSRs) and F2 population-based intra-specific genetic maps in pigeonpea (Bohra et al. 2012). Whereas, the genetic map developed on F2 population derived from Pusa Dwarf’ and ‘HDM04-1’ using 296 genic SNP and SSR markers covered 1520.22 cM (Kumawat et al. 2012). In terms of number of markers mapped in any intra-specific population, the present genetic map has been the most saturated map in pigeonpea as it harbors a total of 787 markers. Few linkage groups in the present genetic map possess a highly dense coverage, while other contained gaps. Inflated length in the present genetic map may be due to segregation distortion observed for a number of SNPs. It is important to mention that GBS has been used to generate SNP genotyping data on the F2 population. GBS seems to be the best option to generate thousands of SNPs in short period of time in any given population. However, reduced representation of genome, problems of missing data, and false-negative genotyping calls especially in detecting heterozygotes in F2 population might have added to the segregation distortion of SNPs generated in the present study. The segregation distortion in pigeonpea F2 populations was observed not only for GBS-based SNPs data, but also for allele-specific polymerase chain reaction (KASPar)-based SNPs data (Saxena et al. 2012) and microsatellite markers (Bohra et al. 2012). The reasons for this segregation distortion could be associated with inherent nature of markers or limitations in genotyping strategy as mentioned above and selection of adaptive genomic regions in breeding cycle of the mapping population development. However, segregation distortion in markers did not exclude them from being potentially informative. In fact, it has been identified that the use of distorted markers could increase the mapping saturation and statistical analysis power for QTL detection (Shizhong 2008a, b). Therefore, in the present study, distorted markers were also included into the linkage analysis.
Mapping data together with DT/IDT phenotyping data on F2 mapping population provided candidate molecular markers and QTLs for growth habit in pigeonpea. The four QTLs detected for growth habit contributed more than 10% of phenotypic variation. These four QTLs have shown contrasting parental effects (three QTLs with positive and one QTL with negative additive effects) including two major QTLs had close map positions on CcLG03; the contrasting parental effects suggested that different QTLs are involved in shaping growth habit in pigeonpea. One QTL from the above-mentioned four QTLs flanked by S3_20698771 and S3_18430894 markers contributing almost two-third of phenotypic variation. This region can be considered as locus for Dt 1 in pigeonpea. One marker S3_20698771 flanking this QTL has been derived from Indel identified in CcTFL1 region. Therefore, location of CcTFL1 in the QTL region further enhances our confidence in defining this region as Dt 1 . These results are in continuation of our previous findings, where we have identified CcTFL1 gene as a potential candidate gene associated with growth habit in pigonpea through sequence polymorphism, candidate gene mapping, and comparative genomics analysis (Mir et al. 2014). Growth habit traits have also been mapped in some other crops species such as soybean, pea, and common bean (Foucher et al. 2003; Kwak et al. 2008; Tian et al. 2010). However, the marker developed by Mir et al. (2014) based on SNP identified (A/T) in CcTFL1 gene was not user friendly; therefore, in the present study, we have analyzed CcTFL1 gene sequence in parental lines of mapping population and detected a 10 bp deletion in DT parent. Functional annotation of identified Indel suggested a frameshift mutation in DT genotype. Furthermore, in comparison with other marker systems, Indels have a number of advantages such as multi-allelic and co-dominant, ease in marker conversion, and most importantly amenable to low-cost genotyping. Indels have been found responsible in altering the gene functions (Jiang et al. 2012). This newly developed Indel-derived marker co-segregated with Dt1 locus in mapping population. Subsequently, this marker has been validated on a set of 262 pigeonpea lines.
The present study in conjunction with Mir et al. (2014) proves that CcTFL1 is a likely candidate gene for growth habit in pigeonpea. The Indel-derived marker based on CcTFL1 will be useful in marker-assisted breeding programs and would allow early generation selection efficiency in crossing programs to select both DT and IDT lines depending on the objective. In the near future, a system level approach to elucidate the gene networks that modulate shoot apical meristem and inflorescence architecture will be required to understand the biological mechanism controlling growth habit in pigeonpea.
Author contribution statement
RKS and RKV conceived, designed, and coordinated the experiments. JO, CVSK, and KBS performed the field experimentations. RKS and JO performed the genotyping. RKS, JO, KBS, and RKV analyzed the data. AS performed the SEM analysis. RKS, JO, AS, and RKV interpreted results. RKS, AS, KBS, and RKV wrote the paper.
References
Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D (2006) A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. The EMBO J 25:605–614
Bahadur B, Rao MM (1981) Seedling handedness in Fabaceae. Proc Indian Acad Sci 90:231–236
Bernard RL (1972) Two genes affecting stem termination in soybean. Crop Sci 12:235–239
Bohra A, Saxena RK, Gnanesh BN, Saxena K, Byregowda M, Rathore A, KaviKishor PB, Cook DR, Varshney RK (2012) An intra-specific consensus genetic map of pigeonpea [Cajanus cajan (L.) Millspaugh] derived from six mapping populations. Theor Appl Genet 125:1325–1338
Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119:721–743
Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E (1996) Control of inflorescence architecture in Antirrhinum. Nature 379:791–797
Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275:80–83
Brimo MC (1983) Inheritance of determinate/indeterminate habit and its affect on yield in broad bean (Vicia faba L.). PhD dissertation, Gottingen Univ, Gottingen, Germany
Elkind Y, Gurnick A, Kedar N (1991) Genetics of semideterminate growth habit in tomato. Hortic Sci 26:1074–1075
Foucher F, Morin J, Courtiade J, Cadioux S, Ellis N, Banfield MJ, Rameau C (2003) DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. Plant Cell 15:2742–2754
Gumber RK, Singh S (1997) Genetics of flowering patterns in pigeonpea: further evidence for two gene control. Euphytica 96:233–236
Gupta SC, Kapoor RK (1991) Inheritance of growth habit in pigeonpea. Crop Sci 31:1456–1459
Gustafson-Brown C, Savidge B, Yanofsky MF (1994) Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell 76:131–143
Hanano S, Goto K (2011) Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23:3172–3184
Hashimoto T (2002) Molecular genetic analysis of left–right handedness in plants. Philos Trans R Soc Lond B Biol Sci 357:799–808
Hegde VS (2011) Morphology and genetics of a new found determinate genotype in chickpea. Euphytica 182:35–42
Jaganathan D, Thudi M, Kale S, Azam S, Roorkiwal M, Gaur PM, Kishor PK, Nguyen H, Sutton T, Varshney RK (2015) Genotyping-by-sequencing based intra-specific genetic map refines a ‘‘QTL-hotspot” region for drought tolerance in chickpea. Mol Genet Genom 290:559–571
Jiang N, Li Z, Wu J, Wang Y, Wu L, Wang S, Wang D, Wen T, Liang Y, Sun P, Liu J (2012) Molecular mapping of the Pi2/9 allelic gene Pi2-2 conferring broad-spectrum resistance to Magnaporthe oryzae in the rice cultivar Jefferson. Rice 5:1
Kempin SA, Savidge B, Yanofsky MF (1995) Molecular basis of the cauliflower phenotype in Arabidopsis. Science 267:522
Kumar V, Khan AW, Saxena RK, Garg V, Varshney RK (2016) First generation hapmap in Cajanus spp. reveals untapped variations in parental lines of mapping populations. Plant Biotechnol J 14:1673–1681
Kumawat G, Raje RS, Bhutani S, Pal JK, Mithra AS, Gaikwad K, Sharma TR, Singh NK (2012) Molecular mapping of QTLs for plant type and earliness traits in pigeonpea (Cajanus cajan L. Millsp.). BMC Genet 13:1
Kwak M, Velasco DM, Gepts P (2008) Mapping homologous sequences for determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris). J Hered 99:283–291
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760
Liu B, Watanabe S, Uchiyama T, Kong F, Kanazawa A, Xia Z, Nagamatsu A, Arai M, Yamada T, Kitamura K, Masuta C, Harada K, Abe J (2010) The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis terminal flower. Plant Physiol 153:198–210
Mandel MA, Yanofsky MF (1995) A gene triggering flower formation in Arabidopsis. Nature 377:522–524
Mir RR, Saxena RK, Saxena KB, Upadhyaya HD, Kilian A, Cook DR, Varshney RK (2012) Whole-genome scanning for mapping determinacy in pigeonpea (Cajanus spp.). Plant Breed 132:472–478
Mir RR, Kudapa H, Srikanth S, Saxena RK, Sharma A, Azam S, Saxena KB, Penmetsa RV, Varshney RK (2014) Candidate gene analysis for determinacy in pigeonpea (Cajanus spp.). Theor Appl Genet 127:2663–2678
Reddy RP, Rao NGP (1974) Inheritance and relation with some yield components of plant and flowering habit in Cajanus. Indian J Genet Plant Breed 34:94–99
Repinski SL, Kwak M, Gepts P (2012) The common bean growth habit gene PvTFL1y is a functional homolog of Arabidopsis TFL1. Theor Appl Genet 124:1539–1547
Royes WV (1976) Pigeonpea. In: Sommonds NW (ed) Evolution of crop plants. Longmans, London and New York, pp 154–156
Ruiz-García L, Madueño F, Wilkinson M, Haughn G, Salinas J, Martínez-Zapater JM (1997) Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9:1921–1934
Sapkal DR, Patil AN, Sapkal VR, Meshram MP (2015) Stem termination genetics in pigeonpea variants (Cajanus cajan (l.) Millsp). The Bioscan 10:1981–1984
Saxena KB, Sharma D (1990) Pigeonpea genetics. In: Nene YL, Hall SD, Sheila VK (eds) The Pigeonpea. CAB International, Wallingford, pp 137–158
Saxena RK, Penmetsa RV, Upadhyaya HD, Kumar A, Carrasquilla-Garcia N, Schlueter JA, Farmer A, Whaley AM, Sarma BK, May GD, Cook DR (2012) Large-scale development of cost-effective single-nucleotide polymorphism marker assays for genetic mapping in pigeonpea and comparative mapping in legumes. DNA Res 19:449–461
Shaw FJC (1936) Studies in Indian pulses. The inheritance of morphological characters and wilt resistance in arhar (Cajanus indicus Spreng.). Indian J Agric Sci 6:139–188
Shizhong X (2008) Quantitative trait locus mapping can benefit from segregation distortion. Genetics 180:2201–2208
Sinjushin AA (2013) Finis coronat axem: terminal inflorescences in tribe Fabeae (Fabaceae: Fabaceae). Wulfenia 20:55–72
Tian Z, Wang X, Lee R, Li Y, Specht JE, Nelson RL, McClean PE, Qiu L, Ma J (2010) Artificial selection for determinate growth habit in soybean. Proc Natl Acad Sci USA 19:8563–8568
Tucker SC (1987) Pseudoracemes in papilionoid legumes: their nature, development, and variation. Bot J Linn Soc 95:181–206
Varshney RK, Chen W, Li Y, Bharti AK, Saxena RK, Schlueter JA, Donoghue MTA, Azam S, Fan G, Whaley AM, Farmer AD, Sheridan J, Iwata A, Tuteja R, Penmetsa RV, Wu W, Upadhyaya HD, Yang S, Shah T, Saxena KB, Michael T, McCombie WR, Yang B, Zhang G, Yang H, Wang J, Spillane C, Cook DR, May GD, Xu X, Jackson SA (2012) Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol 30:83–89
Acknowledgments
Authors are thankful to the United States Agency for International Development (USAID)—India Mission and Department of Agriculture and Cooperation, Ministry of Agriculture & Farmers Welfare, Government of India for the financial support. A.S. acknowledges a partial support from the Russian Foundation for Basic Research (project no. 15-04-06374). Authors would like to thank Abdul Gafoor for technical support. This work has been undertaken as part of the CGIAR Research Program on Grain Legumes. ICRISAT is a member of CGIAR Consortium.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Yunbi Xu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Saxena, R.K., Obala, J., Sinjushin, A. et al. Characterization and mapping of Dt1 locus which co-segregates with CcTFL1 for growth habit in pigeonpea. Theor Appl Genet 130, 1773–1784 (2017). https://doi.org/10.1007/s00122-017-2924-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2924-2