Abstract
Key message
A major QTL for angular leaf spot resistance in the common bean accession G5686 was fine-mapped to a region containing 36 candidate genes. Markers have been developed for marker-assisted selection.
Abstract
Common bean (Phaseolus vulgaris L.) is an important grain legume and an essential protein source for human nutrition in developing countries. Angular leaf spot (ALS) caused by the pathogen Pseudocercospora griseola (Sacc.) Crous and U. Braun is responsible for severe yield losses of up to 80 %. Breeding for resistant cultivars is the most ecological and economical means to control ALS and is particularly important for yield stability in low-input agriculture. Here, we report on a fine-mapping approach of a major quantitative trait locus (QTL) ALS4.1GS, UC for ALS resistance in a mapping population derived from the resistant genotype G5686 and the susceptible cultivar Sprite. 180 F3 individuals of the mapping population were evaluated for ALS resistance and genotyped with 22 markers distributed over 11 genome regions colocating with previously reported QTL for ALS resistance. Multiple QTL analysis identified three QTL regions, including one major QTL on chromosome Pv04 at 43.7 Mbp explaining over 75 % of the observed variation for ALS resistance. Additional evaluation of 153 F4, 89 BC1F2 and 139 F4/F5/BC1F3 descendants with markers in the region of the major QTL delimited the region to 418 kbp harboring 36 candidate genes. Among these, 11 serine/threonine protein kinases arranged in a repetitive array constitute promising candidate genes for controlling ALS resistance. Single nucleotide polymorphism markers cosegregating with the major QTL for ALS resistance have been developed and constitute the basis for marker-assisted introgression of ALS resistance into advanced breeding germplasm of common bean.
Similar content being viewed by others
Introduction
Common bean (Phaseolus vulgaris L.) is the most important grain legume for direct human consumption (Broughton et al. 2003), rich in protein, iron and zinc (Sathe 2002; Hemalatha et al. 2007; Martinez Meyer et al. 2013). The global production of dry and green beans is steadily increasing and has reached 44 million metric tons per year (FAOSTAT 2011). Around 111,000 years ago, common bean diverged from a common ancestor into two different gene pools: large-seeded Andean and small- to medium-seeded Mesoamerican beans (Debouck et al. 1993; Gepts 1998; Singh et al. 2002; Mamidi et al. 2013). Snap beans for green pod harvest are mainly of Andean origin (Gepts and Bliss 1985; Myers and Davis 2002). Common bean is a self-pollinating, diploid species with 11 chromosomes (2n = 2x = 22) (Mok and Mok 1976; Singh 2005), whereof 473 Mbp of DNA sequence have been assembled (Phytozome.net 2014; Schmutz et al. 2014).
Angular leaf spot (ALS) caused by the hemibiotrophic fungus Pseudocercospora griseola (Sacc.) Crous and U. Braun (Bassanezi et al. 2002; Crous et al. 2006) was reported to be responsible for up to 60 and 80 % yield losses in Brazil (de Jesus et al. 2001) and Colombia (Schwartz et al. 1981), respectively. P. griseola spores germinate on the leaf surface after 3 days of moist conditions, enter the leaf through the stomata and grow intercellularly, limited by the leaf veins resulting in an angular lesion shape (Monda et al. 2001; Willocquet et al. 2004). Infection and sporulation occurs in a broad temperature range, from 10 to 33 °C (reviewed in Allorent and Savary 2005). Following the two gene pools of common bean, Mesoamerican and Andean, the pathogen evolved in each gene pool separately (Guzman et al. 1995; Crous et al. 2006). Whereas Andean pathogens are mainly virulent on Andean beans, Mesoamerican and some Andean isolates found in Africa attack both Andeans and Mesoamerican beans (Pastor-Corrales et al. 1998; Mahuku et al. 2002). Since P. griseola is highly variable (Abadio et al. 2012), breeding for a broad ALS resistance involves genes of Andean and Mesoamerican origin or introduction of resistance genes from the secondary gene pool (Mahuku et al. 2003). Genetic resistance is the most efficient strategy to prevent yield loss considering ecological and economic factors (Miklas et al. 2006). However, only few resistance genes have been tagged with closely linked markers.
In Mesoamerican germplasm, the ALS resistance gene Phg-2 was identified in the cultivar Mexico 54 on chromosome Pv08 (Sartorato et al. 2000; Mahuku et al. 2011). Localization of reported markers revealed ALS resistance loci at the same region in Mesoamerican germplasm MAR 2 (Ferreira et al. 2000), Cornell 49-242 (Nietsche et al. 2000), Ouro Negro (Corrêa et al. 2001; Faleiro et al. 2003), G10474 (Mahuku et al. 2004) and G10909 (Mahuku et al. 2011). A genetic test for allelism with one pathotype indicated that the Mesoamerican line BAT 322 contained resistance locus Phg-2 as Mexico 54 (Namayanja et al. 2006), whereas in MAR 2, Mexico 54 and Ouro Negro, five additional independent dominant genes (Phg-3 to Phg-7) were identified using different pathotypes (Caixeta et al. 2003, 2005; Sanglard et al. 2013). As for the Andean lines, several markers linked to ALS resistance have been reported (Table 1). Phg-1 of the Andean cultivar AND 277 (Carvalho et al. 1998) was mapped to chromosome 1 (Goncalves-Vidigal et al. 2011). In the Andean accession G5686, Mahuku et al. (2009) identified a major resistance locus on Pv04 later confirmed by Oblessuc et al. (2012) and named ALS4.1GS, UC. In addition, Mahuku et al. (2009) reported two complementary resistance genes in G5686 on Pv09 (ALS9.1GS) and Pv04 (ALS4.2GS). Further QTL studies support a more quantitative nature of ALS resistance (Lopez et al. 2003; Teixeira et al. 2005; Mahuku et al. 2011; Oblessuc et al. 2012).
Wang et al. (2005) reported a genotyping method based on shifting melting temperatures (T m) of PCR amplicons introduced by allele-specific primers differing in T m. However, the primer design is rather inflexible due to the necessity for the primers to end exactly on the SNP. High-resolution melting curve analysis (HRM) developed by Gundry et al. (2003) and Wittwer et al. (2003) can overcome this disadvantage: Any sequence polymorphism between amplicons can be detected by fluorescence in much larger PCR amplicons (Reed and Wittwer 2004; Montgomery et al. 2007). HRM was used to fine-map resistance loci based on a well-defined DNA sequence polymorphism (Lehmensiek et al. 2008) or even when type, number and composition of the DNA sequence polymorphism in a particular PCR amplicon were unknown (Studer et al. 2009).
In this study, we aimed at (1) validating effective ALS resistance regions in common bean, (2) fine-mapping the source of resistance to identify candidate genes for ALS resistance and (3) developing closely linked markers for breeding applications.
Materials and methods
Plant material
A cross between the resistant Andean common bean genotype G5686 and the susceptible cultivar Sprite had been used for the development of an F2 population to characterize the genetics of ALS resistance (Mahuku et al. 2009). This population was now advanced to an F3 and F4 population with 180 and 153 individuals, respectively. Additionally, a resistant F1 plant backcrossed to the susceptible parental genotype was used to generate a BC1F2 mapping population (89 individuals). G5686 is a highly ALS-resistant Andean dry bean accession from Ecuador (Mahuku et al. 2009). Sprite is an Andean snap bean cultivar (Cunha et al. 2004; Gepts et al. 2008) and was susceptible to over 400 of 503 tested ALS isolates (Mahuku et al. 2009).
Phenotypic evaluation of angular leaf spot (ALS) resistance
Plants were grown in the greenhouse for 17 days at 24–32 °C and infected with the P. griseola isolate 268-COL belonging to pathotype 31-0 (Pastor-Corrales et al. 1998; Mahuku et al. 2009). The inoculation with P. griseola was done according to the CIAT practical guide (Castellanos et al. 2011). The standard visual scale in which 1–3 means resistant, 4–6 intermediate and 7–9 susceptible was used for phenotypic scoring (Schoonhoven and Pastor-Corrales 1987). The plants were evaluated 11, 15 and 20 days after inoculation in the first cycle. Additionally, at the first evaluation, visible hypersensitive reaction (HR) was recorded. In a second infection cycle with 139 selected recombinant descendants, faster disease development was noted. Plants were visually scored after 9, 13 and 15 days post-inoculation. The calculated area under the disease progress curve (AUDPC) was used for classification into susceptible and resistant plants (Mahuku et al. 2009).
Genotyping
Genotyping was conducted according to the strategy of Peleman et al. (2005): The 180 F3 plants were genotyped using two SSR and 17 T m markers which were found to be polymorphic out of a set of 64 tested T m markers (Table 1, Supplemental Table S1). The 153 F4, 89 BC1F2 and 139 F4/F5/BCF3 descendants were genotyped using three selected SNPs (Marker63, Marker50 or Marker9) in order to identify recombination events in a particular region of interest on chromosome Pv04. Finally, 47 recombinant plants were selected and genotyped using three HRM markers, which were found to be polymorphic out of 27 tested amplicons. Primers were designed by the Primer3web version 4.0.0 (Koressaar and Remm 2007; Untergasser et al. 2012).
DNA was extracted according to Xin et al. (2003) from approximately 30 mm2 young trifoliate leaf sample tissue or young unopened trifoliate leaves, using 100 µl buffer A (50 mM NaOH, 2 % Tween 20) and 75 µm buffer B (100 mM Tris–HCl, 1.7 mM EDTA, pH 7.3). The extract was diluted 1:10 in distilled water for PCR.
Simple sequence repeat (SSR) genotyping
SSR markers Pv-ag004 and Pv-ctt001 from Yu et al. (2000) were amplified by PCR in a 20- µl reaction volume. The PCR mix contained 5 μl of genomic DNA solution, 1X Taq buffer [10 mM Tris–HCl pH 8.8, 50 mM KCl, 0.8 % (v/v) Nonidet P40 (Fermentas)], 2.5 mM MgCl2, 0.4 mM dNTPs mix (Promega), 0.2 μM of each primer (forward and reverse) and 0.15 μl of homemade Taq polymerase. 0.1 % Bovine serum albumin (BSA) and 1 % polyvinylpyrrolidone (PVP) were added to counteract polymerase inhibitors present in the DNA extract (Xin et al. 2003).
Pv-ag004 and Pv-ctt001 were amplified under the following PCR conditions: initial denaturation at 94 °C for 3 min followed by 35 cycles of denaturation at 94 °C for 30 s, 48 °C annealing temperature (Yu et al. 2000) for 30 s and extension at 72 °C for 8 min. The SSR marker visualization was described in Mahuku et al. (2009) with a separation at 80 V for approximately 1 h using an Owl T-Rex™ vertical S3S camera (ThermoFisher Scientific Inc, USA).
Melting temperature (T m) shift genotyping
The physical positions of reported markers linked with ALS resistance were identified using sequences from the PhaseolusGenes Toolbox (http://phaseolusgenes.bioinformatics.ucdavis.edu) (2013) and positional information from the Phytozome platform (Goodstein et al. 2012). In those regions (Table 1), SNPs were selected out of a SNP collection provided by The Common Bean Coordinated Agricultural Project (BeanCAP) available on NCBI (2013) and Blair et al. (2013) presenting over 3,300 SNPs. SNP assays were designed following Wang et al. (2005). DNA was amplified by PCR in a total volume of 20 μl containing 4 μl of genomic DNA, 1X Taq buffer, 1.5 mM MgCl2, 0.2 mM dNTPs mix, 0.15 μM each primer (two allele-specific forward primers and the common reverse primer), 1X EvaGreen® (Biotium) and 0.1 μl of homemade Taq polymerase. Amplification was carried out with the following program: initial denaturation at 94 °C for 3 min followed by 35 cycles of denaturation at 92 °C for 15 s, annealing of 15 s (temperature specific to each primer trio, Table 1) and extension at 72 °C for 15 s, finally followed by 10 min extension at 72 °C and 5 min at 10 °C. T m shifts of the amplicons were measured by melting point analysis in a fluorescence-detecting thermocycler (Mx3000P Stratagene) and used to classify the samples into GG (homozygous DNA sequence of G5686), GS (heterozygous) or SS (homozygous DNA sequence of Sprite) genotypes.
Genotyping using high-resolution melting curve analysis (HRM)
HRM genotyping was used to further delimit the position of the QTL ALS4.1GS, UC. Genes of common bean within this region were selected using the Phytozome genome browser (Phytozome.net 2014), and primer pairs spanning introns were designed with the Primer3web version 4.0.0 (Rozen and Skaletsky 2000). For genes without introns, EST sequences of different common bean genotypes were extracted from the database of Ramirez et al. (2005) and CleanEST (Lee and Shin 2009) and aligned to the common bean genome in order to identify SNPs. Primer pairs were designed to flank the SNPs in 150–300 bp distance. Primer sequences were compared against other closely related species such as soybean (Glycine max) or barrel medic (Medicago truncatula) using the Phytozome platform (Goodstein et al. 2012) to select most conserved primer sequences.
Thirteen EST-derived and fourteen intron-flanking primer pairs were designed between 43.5 and 44.5 Mbp on Pv04. The amplification was carried out in a PCR volume of 7.3 μl containing 1X LightScanner high sensitivity master mix (BioFire Diagnostics Inc., UT, USA) including LCGreen® PLUS, 0.10 mM of each forward and reverse primer and 1.3 μl of DNA. Additionally, 14 μl of mineral oil was added to each sample, covering the mix to prevent evaporation during PCR and melting analysis. PCR conditions were set as following: denaturation for 2 min at 95 °C, 40 cycles of 30 s at 94 °C, 30 s annealing at the optimal temperature for each primer pair (Table 1) and 30 s for elongation at 72 °C followed by a final cycle of 2 min at 72 °C, 30 s at 94 °C and 30 s at 25 °C. Using a LightScanner Instrument (BioFire Diagnostics Inc.; 96-well plate format), the amplification product was melted ramping from 60 to 95 °C in 0.05 °C steps per second under continuous fluorescence measurement. The melting curves were related to genotype GG, SS and GS using the LightScanner® and Call-IT® software modules (BioFire Diagnostics Inc.).
Identification of candidate genes within the region of QTL ALS4.1GS, UC
In order to identify homologies to previously reported candidate resistance genes, an NCBI megaBLAST query (Zhang et al. 2000) was conducted using the sequence in the region cosegregating with ALS4.1 of the common bean reference genome (Andean accession G19833, Schmutz et al. 2014). Function of genes analogous to Arabidopsis was studied using The Arabidopsis Information Resource (TAIR) (Swarbreck et al. 2008). Phylogenetic analysis of candidate genes and related common bean genes was carried out using the amino acid sequences of the candidate protein kinases. First, the sequences of the candidate kinases were used for a protein BLAST against the reference genome, collecting the first 100 hits of each candidate sequence. All sequences were then aligned using Clustal Omega (Sievers et al. 2011) and the results analyzed with the software package Mega5 for a comparative analysis and the construction of a maximum-likelihood tree (Tamura et al. 2011).
Statistical analysis
Single and multiple QTL analyses were carried out using the R package qtl developed by Broman et al. (2003). Single QTL analysis and LOD score calculation were done by marker regression and standard interval mapping (200,000 bp steps, 1,000 permutations, 0.01 assumed genotyping error rate) in order to analyze the major QTL. p values were derived by the analysis of variance (ANOVA) if the residues showed normal distribution and using lmPerm R package of Wheeler (2010) if this was not the case. A multiple QTL model was built using the function stepwise with 200,000 bp steps, 1,000 permutations, 0.01 assumed genotyping error rate and 256 imputations in order to calculate a penalized LOD score, followed by analyses omitting one QTL at a time to obtain an ANOVA table.
Results
Phenotypic evaluation for angular leaf spot (ALS) resistance
In order to validate QTL reported in various Andean germplasm, 180 F3 (65 families) of the Sprite × G5686 population were evaluated with the P. griseola pathotype 31-0. Plants were classified into 119 resistant and 61 susceptible plants, showing AUDPC resistance values of <20 and ≥20, respectively. The ratio between the resistant and the susceptible plants is in accordance with one dominant resistance gene segregating in an expected 5:3 ratio, assuming independent F3 individuals (observed Chi-squared = 1.001, p = 0.317). HR was observed in 18 out of the 180 F3 plants. Hypersensitive plants were healthy and vigorous, showing only bright green dots of few millimeters of diameter on the leaf surface without damage to the leaf tissue.
Multiple QTL analysis
Genotyping of these 180 F3 plants with 2 SSR and 20 SNP markers (Table 1) and subsequent multiple QTL analyses identified one major and two minor QTL explaining in total 80.1 % of the phenotypic variation for ALS resistance (Table 2). The major QTL (ALS4.1GS, UC), closely linked to Marker50 localized on chromosome Pv04 at around 43.7 Mbp, explained 75.3 % of the ALS resistance. Two smaller QTL (ALS10.1DG, UC, GS and ALS9.1GS) explained 4.9 and 1.7 % of the ALS resistance, respectively. Out of three markers genotyped on Pv10, Marker17, localized at around 38 Mbp, was best linked to ALS10.1. As for ALS9.1, evaluated with two markers, it was most closely linked to Marker33 localized on Pv09 at around 17.2 Mbp.
Single QTL analysis
Four markers (Marker7, 8, 50 and 9) were analyzed in the region of ALS4.1. Single QTL analysis by interval mapping showed that Marker50 had the most significant effect on ALS resistance (LOD score of 45.9, Fig. 1a), masking all other QTL effects in the single QTL analysis. In accordance with the reported dominant inheritance of ALS resistance, the Marker50 genotypes GG and GS resulted in resistant phenotypes and SS in susceptible phenotypes (permutation p < 2e−16 including 172 observations, Fig. 1a). However, heterozygous GS genotypes had lower resistance levels indicating some codominance effect. Marker50 showed complete linkage with ALS resistance in the sense of all homozygous plants with genotype GG being resistant. Only three plants with a Sprite SS genotype on this locus (according to all four markers in the region) were evaluated as resistant, suggesting phenotypic escapes or involvement of other loci.
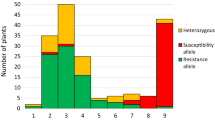
One major QTL controls angular leaf spot (ALS) resistance in the G5686 × Sprite population in an evaluation of 180 F3 plants infected with Pseudocercospora griseola (Sacc.) Crous and U. Braun pathotype (race 31-0). a Box plot sorted by the genotypes at Marker50 shows significant correlation (permutation p < 2e−16, 161 observations) to ALS disease scores, explaining 70.4 % of the phenotypic variation for ALS resistance. The horizontal bar (bold) indicates the median, the box represents inter-quartile range, discontinuous lines represent the upper and lower quartile, and outlier samples (>1.5 × inter-quartile range) are depicted by a circle. Letters indicate significant differences between genotypes using permutation tests. b LOD curve with Marker8, 50 and 9 revealed Marker50 at 43,773,443 bp as closest linked with ALS resistance gene. Dashed line represents LOD score threshold for the 5 % significance level
Marker17, linked to ALS10.1 localized on chromosome Pv010 at around 38.01 Mbp, showed weak linkage with ALS resistance (permutation p < 0.1, 169 observations). Considering only plants with homozygous SS genotypes at Marker50, correlation of Marker17 with ALS resistance was significant (p = 0.003 including 52 observations, Fig. 2a). Marker17 explained 18.2 % of phenotypic variation in those selected plants. Analysis of further markers at the beginning of Pv010, MarkerA1 (p = 0.010, 40 observations) and MarkerA4 (p = 0.044, 40 observations) at 7.15 and 9.73 Mbp, respectively, also resulted in a weak but significant correlation with ALS resistance considering only susceptible SS genotypes at Marker50.
Effects of QTLs ALS10.1DG, UC, GS, ALS9.1GS and ALS5.2UC, GS on angular leaf spot (ALS) resistance in the G5686 × Sprite population infected with Pseudocercospora griseola (Sacc.) Crous and U. Braun pathotype (race 31-0). a Marker17 (ALS10.1) localized on chromosome Pv010 showed significant correlation with ALS resistance (p = 0.003, 52 observations) within the subpopulation of the F3 G5686 × Sprite population with homozygous SS genotypes at Marker50 (ALS4.1GS, UC). b In the same subpopulation, Marker33 (ALS9.1) on Pv09 showed a weak correlation with ALS resistance (p = 0.062, 47 observations). c Marker31 (ALS5.2) localized on Pv05 showed significant influence on ALS resistance (permutation p < 0.05, 130 observations) evaluating the whole F3 population. For each genotype, the horizontal bar (bold) indicates the median, the box represents inter-quartile range, discontinuous lines represent the upper and lower quartile, and outlier samples (>1.5 × inter-quartile range) are depicted by a circle. Letters indicate significant differences between genotypes using TukeyHSD (Marker17) and permutation tests (Marker31)
Marker33 (permutation p < 0.1, 153 observations), linked to ALS9.1 on Pv09, only had a significant effect on ALS resistance in multiple QTL analysis together with Marker32 (permutation p < 0.2, 148 observations), positioned 2.5 Mbp upstream. In the subpopulation considering only plants with SS genotype of Marker50, Marker33 contributed weakly to ALS resistance (p = 0.062, 47 observations, Fig. 2b).
Marker31 (QTL ALS5.2UC, GS) on Pv05 explained 3.7 % of ALS resistance (permutation p < 0.05, 130 observations, Fig. 2c). However, QTL analysis resulted in a LOD score (1.5) below the calculated significance threshold requiring a LOD score above 14.5 according to a 5 % significance level. It was the only marker on Pv05 and could therefore not be included in multiple QTL analysis.
Fine-mapping of the major QTL for ALS resistance
Fine-mapping of the ALS4.1 region was conducted on 47 recombinants out of 561 screened plants (selected from 180 F3, 153 F4, 89 BC1F2 and 139 F4/F5/BC1F3 descendants that were investigated for recombination events in the genomic region of ALS4.1) using seven polymorphic markers: four SNP markers (Marker8, Marker63, Marker50 and Marker9) and three HRM markers (4M437, 4M439 and 4M442). Homozygous plants at 4M437 showed no recombination between the marker and the resistance gene, whereas the flanking Marker63 identified four and 4M439 two recombinant plants (Fig. 3). Hence, the region of ALS4.1 was delimited by the markers Marker63 and 4M439, extending from 43,497,706 to 43,915,434 bp.
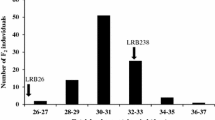
Fine-mapping of QTL ALS4.1GS, UC for angular leaf spot (ALS) resistance in the G5686 × Sprite mapping population points to a locus harboring repetitive serine/threonine protein kinases (STPK). Correlations of Sprite genotypes (SS) and G5686 genotypes (GG) with the susceptible (sus) and resistant (res) phenotypes of selected informative recombinant plants. Analysis of recombinants between flanking Marker63 and 4M439 mapped ALS4.1 to a 417,728-bp region. Shown below, 36 genes within the delimited ALS4.1 region that are annotated in the reference sequence. The delimited ALS4.1 region harbors 11 STPKs containing leucine-rich repeats (Schmutz et al. 2014)
Toward marker-assisted selection
To make this resistance locus available for marker-assisted selection (MAS) to breeding programs, which do not have SNP genotyping facilities, we set up two assays at this locus for outsourcing at the commercial provider LGC genomics (Hertfordshire, UK, http://www.lgcgenomics.com). Assay MAS_ALS4a is based on the polymorphism from Marker50 (Chr04, pos_43,773,443_T/G) and assay MAS_ALS4b (Chr04, pos_43,794,830_T/C) may have a higher specificity, as it was identified based on the recent whole-genome resequencing of genotype G5686 as a unique polymorphism compared with eight other resequenced genotypes (AFR298, G10474, SEA5, MDE23-24, AND696, G40001, G35346 and VAX1, unpublished data). Genotypic data in Table 3 suggest using MAS_ALS4b for marker-assisted selection.
Identification of putative resistance genes within ALS4.1GS, UC
According to the reference sequence of the Andean common bean accession G19833, 36 genes are predicted in the region of the ALS4.1. Resequencing data did not reveal any polymorphisms in the coding sequences of the candidate genes (unpublished data), pointing toward undetected insertions. Among the 36 genes, 11 very closely related to serine/threonine protein kinases (STPK) are annotated (Fig. 3). BLAST searches revealed two Arabidopsis thaliana (At)-like STPKs described as putative resistance genes: Probable LRR receptor-like At1g07560 and At1g18390 involved in response to chitin and respiratory burst defense, best matching common bean genes Phvul.004G154600 and Phvul.004G154800, respectively (Phytozome.net 2014). To further characterize the candidate kinases, the amino acid sequence of Phvul.004G155000 was used for a BLAST search to identify related genes in the bean genome. The amino acid sequences of the first 100 hits were aligned with the candidate STPKs, and a maximum-likelihood tree was built. Eight of the 11 kinases are part of a subfamily, together with other two kinases from Pv02 and 10, and are most related to each other, suggesting that they originate from recent local duplications. The other three kinases were grouped apart and are more closely related to kinases present in Pv07, Pv03 and Pv02. (Supplemental Figure S1). Predicted functions of homologs in plant defense and the repetitive nature make these kinases prime candidates to cause the observed resistance ALS.
Discussion
Characterization of a major resistance locus
Phenotypic evaluation of an F3 mapping population derived from a resistant Andean common bean genotype G5686 and a susceptible cultivar Sprite for ALS resistance demonstrated the presence of one dominant major resistance locus on chromosome Pv04 and three minor loci. Molecular characterization and multiple QTL analyses confirmed a major resistance source (ALS4.1GS, UC) explaining 75.3 % of the phenotypic variation for ALS resistance, which was previously tagged with the SSR marker Pv-ag004 (Mahuku et al. 2009). In the present study, ALS4.1 was delimited to a region between 43,497,706 and 43,915,434 bp on Pv04 and tagged with two markers linked to ALS resistance. The two markers, HRM marker 4M437 and T m shift Marker50, were developed based on EST sequence alignments (Ramirez et al. 2005; Lee and Shin 2009) or SNP collections (NCBI 2013; Blair et al. 2013), respectively. This approach enabled the development of markers at any genomic region, independent of polymorphic microsatellite motifs as necessary for SSR markers. In combination with the HRM technology, allowing for fast and accurate closed-tube genotyping in any genetic background, the markers presented here enable efficient marker-assisted introgression of ALS4.1 into advanced breeding germplasm of common bean. The value of G5686 as an Andean source of resistance to withstand both Andean and Mesoamerican pathotypes of P. griseola has been recognized in previous studies (Pastor-Corrales et al. 1998; Mahuku et al. 2009). Ferreira da Silva et al. (2003) reported marker PV-atct001 linked to a resistant allele in a cross of ESAL 550 and Carioca MG. Lopez et al. (2003) reported a QTL near RGA14 in DOR364 × G19833. Now available physical marker positions suggest these may be the same QTL ALS4.1EC, DG, GS, UC which remains to be confirmed. The ALS resistance gene in ALS4.1 is likely to be one of the genes Phg-2, Phg-3 or Phg-4, reported in AND277 by Caixeta et al. (2005). But as these are not mapped and resistance evaluations used other isolates, further allelism tests are required to identify which gene exactly is underlying ALS4.1
ALS4.1GS, UC embeds a repetitive genome region including potential resistance genes
According to the common bean reference sequence, the ALS4.1 region harbors among its 36 genes repetitive homologs of putative disease resistance genes (Schmutz et al. 2014). Similar observations were reported for other major resistance loci, as, for example, the Mla locus conferring resistance to powdery mildew in barley (Hordeum vulgare L.), where 15 out of 32 annotated protein-coding genes can be associated with plant defense responses (Wei et al. 2002). In the present study, 11 annotated genes containing STPK domains were identified in the region of ALS4.1, eight and three genes, respectively, in two phylogenetically closely related groups. This is interesting since plant resistance genes are often clustered in the genome and evolve rapidly via diverse mechanisms (Smith et al. 2004; David et al. 2009). For example, resistance against soybean cyst nematode is mediated by copy number variation of a 31-kbp genome segment fortifying the expression of several dissimilar genes in a repetitive multigene region (Cook et al. 2012). STPKs phosphorylate hydroxyl groups of serine or threonine residues which is essential for various signaling pathways in eukaryotes including pathogen-triggered immunity (Park et al. 2011; Zhang et al. 2013). For example, the Pto protein in tomato containing a STPK domain (Martin et al. 1993; Loh et al. 1998) induces HR by recognizing two specific Pseudomonas syringae pathogen effectors (Dong et al. 2009). Similar STPK-based defenses were reported in Arabidopsis (Warren et al. 1999; Swiderski and Innes 2001) and wheat against powdery mildew (Cao et al. 2011). The involvement of STPKs in pathogen response and the repetitive arrangement also found at other previously described resistance loci (e.g., Vallejos et al. 2006) make this STPK cluster a likely candidate to cause the observed ALS resistance. Resequencing data of G5686 did not reveal any polymorphisms to the reference G19833 that may cause the resistance. While this may be explained with the difficult assembly of this region due to its various local duplications, we hypothesize the G5686 likely contains additional duplications. To detect new genes, a de novo assembly necessitating deeper sequencing data is required.
Hypersensitive reaction is probably linked with ALS4.1GS, UC and ALS10.1DG, UC, GS
Plants usually respond to biotrophic fungi such as P. griseola at early infection stages with HR (Glazebrook 2005). The At1g18390 gene, sharing 54.3 % protein sequence homology with STPK Phvul.004G154800 in ALS4.1, is likely involved in respiratory burst within the HR (Phytozome.net 2014; TAIR 2013). Indeed, ALS4.1 was linked with observed HR as 16 out of 18 plants (observed χ 2= 5.348, p = 0. 021) showing HR also had GG genotype at Marker50 within ALS4.1. Interestingly, Marker17 linked to ALS10.1 was also associated with HR as 15 out of 17 plants (1 missing data, observed χ 2 = 4.804, p = 0.028) showed both HR and GG genotype at Marker17, possibly explaining the small but significant resistance improvement when both QTL present. This is in line with Zhou et al. (1995), who observed enhanced HR in the presence of additional genes besides resistance genes Pto and Prf. The fact that HR was observed only in 18 out of 180 plants of the F3 population indicates that HR may require several genes (Zhou et al. 1995; Salmeron et al. 1996), a low phenotypic penetrance, or that activation of systemic acquired resistance (Oh and Martin 2011) prevented the plants from visible symptoms.
ALS4.1GS, UC is a major QTL suitable for marker-assisted introgression of ALS resistance
In a previous study, three dominant and complementary genes linked with SSR markers Pv-ag004 (ALS4.1 region), Pv-at007 (ALS10.1 region) and Pv-ctt001 (beginning of Pv04), respectively, were reported to confer ALS resistance in an F2 population of the same cross infected with the same P. griseola pathotype (Mahuku et al. 2009). The authors concluded the involvement of three dominant and complementary genes, statistically expecting 56.3 % of plants which carry a G allele at Pv-ag004 (ALS4.1) to be resistant to ALS, whereas the observed proportion was 68 %. In contrast, in the current study, 89 % of plants with a G allele at Pv-ag004 were resistant to ALS, and even 97 % of the plants with the homozygous genotype GG (data not shown). These percentages depend on the classification of heterozygous plants to be resistant or susceptible, which might have been different in Mahuku et al. (2009) and confounded the effect of Pv-ag004. Moreover, environmental conditions might have affected the otherwise identical phenotyping procedure. In the current study, ALS4.1 was masking the minor effect of Pv-at007 linked to ALS9.1GS and an effect of Pv-ctt001 could not be detected (permutation p > 0.5 including 78 observations). Consistent detection of ALS4.1 across repeated experiments explaining the major proportion of the observed resistance rather indicates that ALS resistance in this population is controlled by one independent major locus modified by few minor QTL. We conclude that ALS4.1 has a high value as a source for marker-assisted introgression of ALS resistance, with polymorphism T/C on Chr04 at 43,794,830 bp (marker MAS_ALS4b) showing highest specificity for G5686.
Various QTL control ALS resistance
ALS resistance loci in the region of ALS4.1 were reported in different germplasm screened with several pathotypes (Faleiro et al. 2003; Lopez et al. 2003; Caixeta et al. 2005; Mahuku et al. 2011) and in different environments (Oblessuc et al. 2012). For example, Ferreira da Silva et al. (2003) identified a major ALS resistance locus in the same region as ALS4.1 evaluating the ESAL 550 cultivar in a field experiment. Lopez et al. (2003) and Oblessuc et al. (2012) reported QTL in the same region explaining a smaller part of the variation using various Andean genotypes and experimental conditions. Oblessuc et al. (2012) named two closely adjacent QTL on Pv04, ALS4.1 and ALS4.2, which encase the QTL reported here. Hence, we hypothesize that these are actually one QTL named ALS4.1GS, UC. AND277, the only genotype to share the G5686 allele for the MAS_ALS4b marker in the evaluated set (Table 1), is similar but not identical to G5686 (correlation 0.88 based on 650 markers, unpublished data). Also available pedigree information does not indicate G5686 in the ancestry of AND277; hence, there is no clear evidence that G5686 and AND277 carry the same allele. On Pv10, Oblessuc et al. (2012) identified the major QTL ALS10.1DG, UC, GS (Marker17) explaining over 20 % of ALS resistance in the field, in both dry and wet season using CAL 143 as a source of resistance (Oblessuc et al. 2012). ALS9.1GS was previously described by Mahuku et al. (2009) but not named. Taken together, the significant QTL in this study was also found in other experiments and genotypes, supporting the findings in this work.
ALS resistance initially reported to be monogenetic evaluating only one pathotype (Ferreira et al. 2000; Corrêa et al. 2001) was shown to be quantitatively inherited evaluating different pathotypes (Faleiro et al. 2003; Caixeta et al. 2005). Pathotype-specific resistance in common bean was also reported considering different pathotypes of rust (Park et al. 1999; Faleiro et al. 2003) and anthracnose (Faleiro et al. 2003; Rodriguez-Suarez et al. 2007). Since G5686 proved resistance against a wide range of pathotypes (Mahuku et al. 2009), it appears likely that minor QTL or additional QTL will add specific resistance to other pathotypes.
Conclusion
A major QTL explaining 75.3 % of ALS resistance in the G5686 × Sprite population was validated, mapped to 418 kbp on chromosome Pv04 and tagged with two closely linked SNP markers (Marker50 and 4M437) allowing efficient MAS. ALS4.1GS, UC defines a region of 36 genes including 11 STPKs, which are likely candidates for the resistance gene. Additionally, three minor QTLs were identified. The Andean resistance loci ALS4.1 and Phg-1 as well as the Mesoamerican Phg-2 can now be combined and tested in elite cultivars in order to pyramid resistance genes.
Author contribution statement
BR, CJ, CM, BS and BK conceived and designed the study, and BK performed the experiments. BR, CJ and BS contributed with material, reagents and analysis tools, while BK, BR, JDL and CM helped analyzing the data. The manuscript was drafted by BK and further improved by CM, BR and BS. All authors read and approved the manuscript for publication.
References
Abadio AKR, Lima SS, Santana MF, Salomao TMF, Sartorato A, Mizubuti ESG, Araujo EF, de Queiroz MV (2012) Genetic diversity analysis of isolates of the fungal bean pathogen Pseudocercospora griseola from central and southern Brazil. Genet Mol Res 11(2):1272–1279. doi:10.4238/2012.May.14.1
Allorent D, Savary S (2005) Epidemiological characteristics of angular leaf spot of bean: a systems analysis. Eur J Plant Pathol 113(4):329–341
Bassanezi RB, Amorim L, Filho AB, Berger RD (2002) Gas exchange and emission of chlorophyll fluorescence during the monocycle of rust, angular leaf spot and anthracnose on bean leaves as a function of their trophic characteristics. J Phytopathol 150(1):37–47
Blair MW, Cortes AJ, Penmetsa RV, Farmer A, Carrasquilla-Garcia N, Cook DR (2013) A high-throughput SNP marker system for parental polymorphism screening, and diversity analysis in common bean (Phaseolus vulgaris L.). Theor Appl Genet 126(2):535–548. doi:10.1007/s00122-012-1999-z
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19(7):889–890. doi:10.1093/bioinformatics/btg112
Broughton WJ, Hernandez G, Blair M, Beebe S, Gepts P, Vanderleyden J (2003) Beans (Phaseolus spp.)—model food legumes. Plant Soil 252(1):55–128. doi:10.1023/a:1024146710611
Caixeta ET, Borem A, Fagundes SD, Niestche S, de Barros EG, Moreira MA (2003) Inheritance of angular leaf spot resistance in common bean line BAT 332 and identification of RAPD markers linked to the resistance gene. Euphytica 134(3):297–303
Caixeta ET, Borem A, Alzate-Marin AL, Fagundes SD, Silva M, de Barros EG, Moreira MA (2005) Allelic relationships for genes that confer resistance to angular leaf spot in common bean. Euphytica 145(3):237–245. doi:10.1007/s10681-005-1258-3
Cao A, Xing L, Wang X, Yang X, Wang W, Sun Y, Qian C, Ni J, Chen Y, Liu D, Wang X, Chen P (2011) Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc Natl Acad Sci. doi:10.1073/pnas.1016981108
Carvalho GA, Paula Junior TJ, Alzate-Marin AL, Nietsche S, Barros EG, Moreira MA (1998) Inheritance of resistance of the Andean bean line AND-277 to race 63-23 of Phaeoisariopsis griseola and identification of a RAPD marker linked to the resistance gene. Fitopatol Bras 23(4):482–485
Castellanos G, Jara C, Mosquera G (2011) Manejo del hongo en el laboratorio. Phaeoisariopsis griseola, vol 6. Centro Internacional de Agricultura Tropical (CIAT), Cali-Palmira
Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, Wang J, Hughes TJ, Willis DK, Clemente TE, Diers BW, Jiang J, Hudson ME, Bent AF (2012) Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338(6111):1206–1209. doi:10.1126/science.1228746
Corrêa R, Good-God P, Oliveira M, Niestche S, Moreira M, de Barros E (2001) Herança da resistência á manchaangular do feijoeiro e identificaçao de marcadores moleculares flanqueando o loco de resistência. Fitopatol Bras 26:7–32
Crous PW, Liebenberg MM, Braun U, Groenewald JZ (2006) Re-evaluating the taxonomic status of Phaeoisariopsis griseola, the causal agent of angular leaf spot of bean. Stud Mycol 55:163–173
Cunha C, Hintz T, Griffiths P (2004) Genetic diversity of snap bean cultivars determined using randomly amplified polymorphic DNA (RAPD) markers. HortScience 39(3):481–484
David P, Chen NWG, Pedrosa-Harand A, Thareau V, Sevignac M, Cannon SB, Debouck D, Langin T, Geffroy V (2009) A nomadic subtelomeric disease resistance gene cluster in common bean. Plant Physiol 151(3):1048–1065. doi:10.1104/pp.109.142109
Debouck DG, Toro O, Paredes OM, Johnson WC, Gepts P (1993) Genetic diversity and ecological distribution of Phaseolus vulgaris (Fabaceae) in northwestern South America. Econ Bot 47(4):408–423. doi:10.1007/bf02907356
de Jesus WC, do Vale FX, Coelho RR, Hau B, Zambolim L, Costa LC, Filho AB (2001) Effects of angular leaf spot and rust on yield loss of phaseolus vulgaris. Phytopathology 91(11):1045–1053
Dong J, Xiao F, Fan F, Gu L, Cang H, Martin GB, Chai J (2009) Crystal structure of the complex between pseudomonas effector AvrPtoB and the tomato Pto kinase reveals both a shared and a unique interface compared with AvrPto-Pto. Plant Cell 21(6):1846–1859. doi:10.1105/tpc.109.066878
Faleiro F, Ragagnin V, Schuster I, Corrêa R, Good-God P, Brommonshenkel S, Moreira M, Barros E (2003) Mapeamento de genes de resistência do feijoeiro à ferrugem, antracnose e mancha-angular usando marcadores RAPD. Fitopatol Bras 28:59–66
FAOSTAT (2011) FAO statistic. Food and agriculture organization of the united nations (FAO). http://faostat.fao.org/. Accessed 16 May 2013
Ferreira da Silva G, Bosco dos Santos J, Patto Ramalho MA (2003) Identification of SSR and RAPD markers linked to a resistance allele for angular leaf spot in the common bean (Phaseolus vulgaris) line ESAL 550. Genetics and Molecular Biology 26(4):459–463
Ferreira CF, Borem A, Carvalho GA, Nietsche S, Paula TJ, Barros EG, Moreira MA (2000) Inheritance of angular leaf spot resistance in common bean and identification of a RAPD marker linked to a resistance gene. Crop Sci 40(4):1130–1133
Gepts P (1998) Origin and evolution of common bean: past events and recent trends. HortScience 33(7):1124–1130
Gepts P, Bliss FA (1985) F1 hybrid weakness in the common bean; differential geographic origin suggests two gene pools in cultivated bean germplasm. J Hered 76:447–450
Gepts P, Aragao FJL, de Barros E, Blair MW, Brondani R, Broughton W, Galasso I, Hernandez G, Kami J, Lariguet P, McClean P, Melotto M, Miklas P, Pauls P, Pedrosa-Harand A, Porch T, Sanchez F, Sparvoli F, Yu K (2008) Genomics of Phaseolus beans, a major source of dietary protein and micronutrients in the tropics. In: Moore PH, Ming R (eds) Genomics of tropical crop plants, vol 1. Plant Genetics and Genomics-Crops and Models, Springer, New York, pp 113–143
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227. doi:10.1146/annurev.phyto.43.040204.135923
Goncalves-Vidigal MC, Cruz AS, Garcia A, Kami J, Vidigal Filho PS, Sousa LL, McClean P, Gepts P, Pastor-Corrales MA (2011) Linkage mapping of the Phg-1 and Co-1 (4) genes for resistance to angular leaf spot and anthracnose in the common bean cultivar AND 277. Theor Appl Genet 122(5):893–903. doi:10.1007/s00122-010-1496-1
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40(D1):D1178–D1186. doi:10.1093/nar/gkr944
Gundry CN, Vandersteen JG, Reed GH, Pryor RJ, Chen J, Wittwer CT (2003) Amplicon melting analysis with labeled primers: A closed-tube method for differentiating homozygotes and heterozygotes. Clin Chem 49(3):396–406. doi:10.1373/49.3.396
Guzman P, Gilbertson RL, Nodari R, Johnson WC, Temple SR, Mandala D, Mkandawire ABC, Gepts P (1995) Characterization of variability in the fungus Phaeoisariopsis griseola suggests coevolution with the common bean (Phaseolus vulgaris). Phytopathology 85(5):600–607. doi:10.1094/Phyto-85-600
Hemalatha S, Platel K, Srinivasan K (2007) Zinc and iron contents and their bioaccessibility in cereals and pulses consumed in India. Food Chem 102(4):1328–1336. doi:10.1016/j.foodchem.2006.07.015
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23(10):1289–1291. doi:10.1093/bioinformatics/btm091
Lee B, Shin G (2009) CleanEST: a database of cleansed EST libraries. Nucleic Acids Res 37:D686–D689. doi:10.1093/nar/gkn648
Lehmensiek A, Sutherland MW, McNamara RB (2008) The use of high resolution melting (HRM) to map single nucleotide polymorphism markers linked to a covered smut resistance gene in barley. Theor Appl Genet 117(5):721–728. doi:10.1007/s00122-008-0813-4
Loh YT, Zhou JM, Martin GB (1998) The myristylation motif of Pto is not required for disease resistance. Mol Plant Microbe Interact 11(6):572–576. doi:10.1094/mpmi.1998.11.6.572
Lopez CE, Acosta IF, Jara C, Pedraza F, Gaitan-Solis E, Gallego G, Beebe S, Tohme J (2003) Identifying resistance gene analogs associated with resistances to different pathogens in common bean. Phytopathology 93(1):88–95. doi:10.1094/phyto.2003.93.1.88
Mahuku GS, Henriquez MA, Munoz J, Buruchara RA (2002) Molecular markers dispute the existence of the Afro-Andean group of the bean angular leaf spot pathogen, Phaeoisariopsis griseola. Phytopathology 92(6):580–589. doi:10.1094/phyto.2002.92.6.580
Mahuku GS, Jara C, Cajiao E, Beebe S (2003) Sources of resistance to angular leaf spot (Phaeoisariopsis griseola) in common bean core collection, wild Phaseolus vulgaris and secondary gene pool. Euphytica 130(3):303–313. doi:10.1023/a:1023095531683
Mahuku G, Montoya C, Henriquez MA, Jara C, Teran H, Beebe S (2004) Inheritance and characterization of angular leaf spot resistance gene present in common bean accession G 10474 and identification of an AFLP marker linked to the resistance gene. Crop Sci 44(5):1817–1824
Mahuku GS, Maria Iglesias A, Jara C (2009) Genetics of angular leaf spot resistance in the Andean common bean accession G5686 and identification of markers linked to the resistance genes. Euphytica 167(3):381–396. doi:10.1007/s10681-009-9897-4
Mahuku GS, Antonia Henriquez M, Montoya C, Jara C, Teran H, Beebe S (2011) Inheritance and development of molecular markers linked to angular leaf spot resistance genes in the common bean accession G10909. Mol Breed 28(1):57–71. doi:10.1007/s11032-010-9461-x
Mamidi S, Rossi M, Moghaddam SM, Annam D, Lee R, Papa R, McClean PE (2013) Demographic factors shaped diversity in the two gene pools of wild common bean Phaseolus vulgaris L. Heredity 110(3):267–276. doi:10.1038/hdy.2012.82
Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu TY, Earle ED, Tanksley SD (1993) Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262(5138):1432–1436. doi:10.1126/science.7902614
Martinez Meyer MR, Rojas A, Santanen A, Stoddard FL (2013) Content of zinc, iron and their absorption inhibitors in Nicaraguan common beans (Phaseolus vulgaris L.). Food Chem 136 (1):87–93
Miklas PN, Kelly JD, Beebe SE, Blair MW (2006) Common bean breeding for resistance against biotic and abiotic stresses: from classical to MAS breeding. Euphytica 147(1–2):105–131. doi:10.1007/s10681-006-4600-5
Mok DWS, Mok MC (1976) A modified Giemsa technique for identifying bean chromosomes. J Hered 67(3):187–188
Monda EO, Sanders FE, Hick A (2001) Infection and colonization of bean leaf by Phaeoisariopsis griseola. Plant Pathol 50(1):103–110. doi:10.1046/j.1365-3059.2001.00537.x
Montgomery J, Wittwer CT, Palais R, Zhou LM (2007) Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis. Nat Protoc 2(1):59–66. doi:10.1038/nprot.2007.10
Myers J, Davis J (2002) Molecular polygenetics of snap bean. Department of Horticulture, Oregon State University, Corvallis
Namayanja A, Buruchara R, Mahuku G, Rubaihayo P, Kimani P, Mayanja S, Eyedu H (2006) Inheritance of resistance to angular leaf spot in common bean and validation of the utility of resistance linked markers for marker assisted selection out side the mapping population. Euphytica 151(3):361–369. doi:10.1007/s10681-006-9158-8
NCBI (2013) GenBank. National Center for Biotechnology Information (NCBI). http://www.ncbi.nlm.nih.gov/genbank. Accessed 11 July 2013
Nietsche S, Borem A, Carvalho GA, Rocha RC, Paula TJ, de Barros EG, Moreira MA (2000) RAPD and SCAR markers linked to a gene conferring resistance to angular leaf spot in common bean. J Phytopathol-Phytopathol Z 148(2):117–121
Oblessuc PR, Baroni RM, Garcia AAF, Chioratto AF, Carbonell SAM, Camargo LEA, Benchimol LL (2012) Mapping of angular leaf spot resistance QTL in common bean (Phaseolus vulgaris L.) under different environments. BMC Genet 13(50):9. doi:10.1186/1471-2156-13-50
Oblessuc PR, Perseguini J, Baroni RM, Chiorato AF, Carbonell SAM, Mondego JMC, Vidal RO, Camargo LEA, Benchimol-Reis LL (2013) Increasing the density of markers around a major QTL controlling resistance to angular leaf spot in common bean. Theor Appl Genet 126(10):2451–2465. doi:10.1007/s00122-013-2146-1
Oh CS, Martin GB (2011) Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci 16(3):132–140. doi:10.1016/j.tplants.2010.11.001
Park SO, Coyne DP, Bokosi JM, Steadman JR (1999) Molecular markers linked to genes for specific rust resistance and indeterminate growth habit in common bean. Euphytica 105(2):133–141. doi:10.1023/a:1003477714349
Park G, Servin JA, Turner GE, Altamirano L, Colot HV, Collopy P, Litvinkova L, Li LD, Jones CA, Diala FG, Dunlap JC, Borkovich KA (2011) Global analysis of serine-threonine protein kinase genes in Neurospora crassa. Eukaryot Cell 10(11):1553–1564. doi:10.1128/ec.05140-11
Pastor-Corrales MA, Jara C, Singh SP (1998) Pathogenic variation in, sources of, and breeding for resistance to Phaeoisariopsis griseola causing angular leaf spot in common bean. Euphytica 103(2):161–171. doi:10.1023/a:1018350826591
Peleman JD, Wye C, Zethof J, Sorensen AP, Verbakel H, van Oeveren J, Gerats T, van der Voort JR (2005) Quantitative trait locus (QTL) isogenic recombinant analysis: a method for high-resolution mapping of QTL within a single population. Genetics 171(3):1341–1352. doi:10.1534/genetics.105.045963
PhaseolusGenes (2013) Bean Breeder’s molecular marker toolbox. UC Davis Bioinformatics Core at University of California, Davis. http://phaseolusgenes.bioinformatics.ucdavis.edu. Accessed 23 May 2013
Phytozome.net (2014) Phaseolus vulgaris v1.0. DOE-JGI and USDA-NIFA. http://www.phytozome.net/. Accessed 20 May 2014
Queiroz VT, Sousa CS, Costa MR, Sanglad DA, Arruda KMA, Souza TLPO, Ragagnin VA, Barros EG, Moreira MA (2004) Development of SCAR markers linked to common bean angular leaf spot resistance genes. Annu Rep Bean Improv Coop 47:237–238
Ramirez M, Graham MA, Blanco-Lopez L, Silvente S, Medrano-Soto A, Blair MW, Hernandez G, Vance CP, Lara M (2005) Sequencing and analysis of common bean ESTs. Building a foundation for functional genomics. Plant Physiol 137(4):1211–1227. doi:10.1104/pp.104.054999
Reed GH, Wittwer CT (2004) Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem 50(10):1748–1754. doi:10.1373/clinchem.2003.029751
Rodriguez-Suarez C, Mendez-Vigo B, Paneda A, Ferreira JJ, Giraldez R (2007) A genetic linkage map of Phaseolus vulgaris L. and localization of genes for specific resistance to six races of anthracnose (Colletotrichum lindemuthianum). Theor Appl Genet 114(4):713–722. doi:10.1007/s00122-006-0471-3
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology, Humana Press, Totowa
Salmeron JM, Oldroyd GED, Rommens CMT, Scofield SR, Kim HS, Lavelle DT, Dahlbeck D, Staskawicz BJ (1996) Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86(1):123–133. doi:10.1016/s0092-8674(00)80083-5
Sanglard DA, Gomes Ribeiro CA, Pereira Balbi B, Antunes Arruda KM, Gonçalves de Barros E, Alves Moreira M (2013) Characterization of the angular leaf spot resistance gene present in common bean cultivar Ouro Negro. J Agric Sci 5(2):19–23
Sartorato A, Nietsche S, Barros EG, Moreira MA (2000) RAPD and SCAR markers linked to resistance gene to angular leaf spot in common beans. Fitopatol Bras 25:637–642
Sathe SK (2002) Dry bean protein functionality. Crit Rev Biotechnol 22(2):175–223
Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB, Grimwood J, Jenkins J, Shu S, Song Q, Chavarro C, Torres-Torres M, Geffroy V, Moghaddam SM, Gao D, Abernathy B, Barry K, Blair M, Brick MA, Chovatia M, Gepts P, Goodstein DM, Gonzales M, Hellsten U, Hyten DL, Jia G, Kelly JD, Kudrna D, Lee R, Richard MMS, Miklas PN, Osorno JM, Rodrigues J, Thareau V, Urrea CA, Wang M, Yu Y, Zhang M, Wing RA, Cregan PB, Rokhsar DS, Jackson SA (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46:707–713
Schoonhoven AV, Pastor-Corrales MA (1987) Standard system for the evaluation of bean germplasm. Centro Internacional de Agricultura Tropical, Cali
Schwartz HF, Correa VF, Pineda DPA, Otoya MM, Katherman MJ (1981) Dry bean yield losses caused by Ascochyta, angular, and white leaf spots in Colombia. Plant Dis 65:494–496
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soeding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7(539):6. doi:10.1038/msb.2011.75
Singh SP (2005) Common bean (Phaseolus vulgaris L.). In: Singh RJ, Jauhar PP (eds) Genetic resources, chromosome engineering, and crop improvement: grain legumes, vol 1. CRC Press, USA
Singh SP, Terán H, Muñoz CG, Osorno JM (2002) Selection for seed yield in Andean intra-gene pool and Andean×Middle American inter-gene pool populations of common bean. Euphytica 127(3):437–444. doi:10.1023/A:1020317608553
Smith SM, Pryor AJ, Hulbert SH (2004) Allelic and haplotypic diversity at the Rp1 rust resistance locus of maize. Genetics 167(4):1939–1947. doi:10.1534/genetics.104.029371
Studer B, Jensen LB, Fiil A, Asp T (2009) “Blind” mapping of genic DNA sequence polymorphisms in Lolium perenne L. by high resolution melting curve analysis. Mol Breed 24(2):191–199. doi:10.1007/s11032-009-9291-x
Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, Radenbaugh A, Singh S, Swing V, Tissier C, Zhang P, Huala E (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36:D1009–D1014. doi:10.1093/nar/gkm965
Swiderski MR, Innes RW (2001) The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J Cell Mol Biol 26(1):101–112
TAIR (2013) The Arabidopsis information resource (TAIR). http://arabidopsis.org. Accessed 11 July 2013
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. doi:10.1093/molbev/msr121
Teixeira FF, Bosco dos Santos J, Patto Ramalho MA, Barbosa Abreu ÂdF, Teixeira Guimarães C, Carlos de Oliveira A (2005) QTL mapping for angular leaf spot in common bean using microsatellite markers. Crop Breed Appl Biotechnol 5:272–278
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3–new capabilities and interfaces. Nucl Acids Res 40(15):e115. doi:10.1093/nar/gks596
Vallejos EC, Astua-Monge G, Jones V, Plyler TR, Sakiyama NS, Mackenzie SA (2006) Genetic and molecular characterization of the I locus of Phaseolus vulgaris. Genetics 172(2):1229–1242
Wang J, Chuang K, Ahluwalia M, Patel S, Umblas N, Mirel D, Higuchi R, Germer S (2005) High-throughput SNP genotyping by single-tube PCR with T-m-shift primers. Biotechniques 39(6):885–893. doi:10.2144/000112028
Warren RF, Merritt PM, Holub E, Innes RW (1999) Identification of three putative signal transduction genes involved in R gene-specified disease resistance in Arabidopsis. Genetics 152(1):401–412
Wei FS, Wong RA, Wise RP (2002) Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14(8):1903–1917. doi:10.1105/tpc.002238
Wheeler B (2010) lmPerm: permutation tests for linear models, R package. http://www.icesi.edu.co/CRAN/src/contrib/Archive/lmPerm/lmPerm_1.1-0.tar.gz
Willocquet L, Allorent D, Savary S (2004) Quantitative analysis of two important epidemiological features of the common bean: Phaeoisariopsis griseola pathosystem. Fitopatol Bras 29(6):676–679
Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ (2003) High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem 49(6):853–860. doi:10.1373/49.6.853
Xin ZG, Velten JP, Oliver MJ, Burke JJ (2003) High-throughput DNA extraction method suitable for PCR. Biotechniques 34(4):820–826
Yu K, Park SJ, Poysa V, Gepts P (2000) Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.). J Hered 91(6):429–434. doi:10.1093/jhered/91.6.429
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7(1–2):203–214. doi:10.1089/10665270050081478
Zhang Y, Lubberstedt T, Xu ML (2013) The genetic and molecular basis of plant resistance to pathogens. J Genet Genom 40(1):23–35. doi:10.1016/j.jgg.2012.11.003
Zhou J, Loh Y-T, Bressan RA, Martin GB (1995) The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 83(6):925–935. doi:10.1016/0092-8674(95)90208-2
Acknowledgments
The authors would like to acknowledge the colleagues at CIAT for greenhouse operations and support in the laboratory. We sincerely thank Prof. Dr. Achim Walter for hosting the Forage Crop Genetics group at ETH Zurich. This study was partly funded by the SNSF Professorship (Grant No: PP00P2_138988) and GCP, Improving tropical legume productivity for marginal environments in sub-Saharan Africa (Grant No: OPPGD 1392).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The experiments shown in the manuscripts submitted for publication comply with the current laws of the country in which they were performed.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. A. Lightfoot.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2015_2472_MOESM1_ESM.pdf
Figure S1 Phylogenetic analysis of common bean kinases most related to those found at QTL ALS4.1GS, UC. BLAST searches of all 11 serine/threonine protein kinases (STPK) within ALS4.1, marked in yellow, were carried out to identify most related STPKs in the bean reference genome (Schmutz et al. 2014). STPKs in ALS4.1 fall into two groups of highly related genes, indicating local duplications (PDF 22 kb)
122_2015_2472_MOESM2_ESM.xlsx
Table S1 Primer sequences Names of polymorphic markers, single nucleotide polymorphism (SNP) ID, sequences and physical position of all used primers are listed. (XLSX 23 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Keller, B., Manzanares, C., Jara, C. et al. Fine-mapping of a major QTL controlling angular leaf spot resistance in common bean (Phaseolus vulgaris L.). Theor Appl Genet 128, 813–826 (2015). https://doi.org/10.1007/s00122-015-2472-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2472-6