Abstract
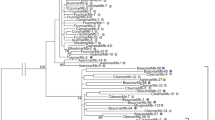
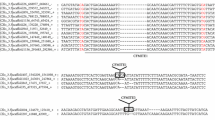
Three different complete mariner elements were found in the genome of the ant Tapinoma nigerrimum. One (Tnigmar-Mr) was interrupted by a 900-bp insertion that corresponded to an incomplete member of a fourth mariner element, called Azteca. In this work, we isolate and characterize full-length Tnigmar-Az elements in T. nigerrimum. The purpose of this study is to clarify the evolutionary history of Azteca elements and their hosts as well as the possible existence of horizontal transfer processes. For this, Azteca-like elements were also retrieved from the available sequences of various ant genomes, representing four different ant subfamilies: Dolichoderinae, Formicinae, Myrmicinae, and Ponerinae. The tree topology resulting for the Azteca-like elements bore very little resemblance to that of their respective hosts. The pervasive presence of Azteca-like elements in all ant genomes, together with the observation that extant copies are usually younger than the genomes that host them, could be explained either by lineage sorting or by recent horizontal transfer of active elements. However, the finding of closer genetic relationships between elements than between the ants that host them is consistent with the latter scenario. This is clearly observed in Sinvmar-Az, Tnigmar-Az, Acepmar-Az, and Cflomar-Az elements, suggesting the existence of horizontal transfer processes. On the contrary, some elements displayed more divergence than did the hosts harboring them. This may reflect either further horizontal transfer events or random lineage sorting.



Similar content being viewed by others
References
Aris-Brosou S (2003) Least and most powerful phylogenetic tests to elucidate the origin of the seed plants in the presence of conflicting signals under misspecified models. Syst Biol 52:781–793
Assimacopoulos S, Charlesworth B (2005) Fixation of transposable elements in the Drosophila melanogaster genome. Genet Res 85:195–203
Barry EG, Witherspoon DJ, Lampe DJ (2004) A bacterial genetic screen identifies functional coding sequences of the insect mariner transposable element Famar1 amplified from the genome of the earwig, Forficula auricularia. Genetics 166:823–833
Bartolomé C, Maside X (2004) The lack of recombination drives the fixation of transposable elements on the fourth chromosome of Drosophila melanogaster. Genet Res 83:91–100
Bartolomé C, Maside X, Charlesworth B (2002) On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol Biol Evol 19:926–937
Bartolomé C, Bello X, Maside X (2009) Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol 10:R22
Bennetzen JL, Wang H (2014) The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu Rev Plant Biol 65:505–530
Bigot Y, Hamelin MH, Capy P, Periquet G (1994) Mariner-like elements in hymenopteran species, Insertion site and distribution. Proc Natl Acad Sci U S A 91:3408–3412
Bonasio R, Zhang G, Ye C, Mutti NS, Fang X, Qin N, Donahue G, Yang P, Li Q, Li C, Zhang P, Huang Z, Berger SL, Reinberg D, Wang J, Liebig J (2010) Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329:1068–1071
Bouuaert C, Chalmers R (2010) Transposition of the human Hsmar1 transposon: rate-limiting steps and the importance of the flanking TA dinucleotide in second strand cleavage. Nucleic Acids Res 38:190–202
Brady SG, Schultz TR, Fisher BL, Ward PS (2006) Evaluating alternative hypotheses for the early evolution and diversification of ants. Proc Natl Acad Sci U S A 103:18172–18177
Bui QT, Casse N, Leignel V, Nicolas V, Chénais B (2008) Widespread occurrence of mariner transposons in coastal crabs. Mol Phylogenet Evol 47:1181–1189
Cuypers MG, Trubitsyna M, Callow P, Forsyth VT, Richardson JM (2013) Solution conformations of early intermediates in Mos1 transposition. Nucleic Acids Res 41:2020–2033
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Gadau J, Helmkampf M, Nygaard S, Roux J, Simola DF, Smith CR, Suen G, Wurm Y, Smith CD (2012) The genomic impact of 100 million years of social evolution in seven ant species. Trends Genet 28:14–28
Garcia Guerreiro MP, Fontdevila A (2007) The evolutionary history of Drosophila buzzatii. XXXVI. Molecular structural analysis of Osvaldo retrotransposon insertions in colonizing populations unveils drift effects in founder events. Genetics 175:301–310
González J, Petrov DA (2009) The adaptive role of transposable elements in the Drosophila genome. Gene 448:124–133
González J, Macpherson JM, Petrov DA (2009) A recent adaptive transposable element insertion near highly conserved developmental loci in Drosophila melanogaster. Mol Biol Evol 26:1949–1961
Hartl DL (2001) Discovery of the transposable element mariner. Genetics 157:471–476
Iranzo J, Gómez MJ, López de Saro FJ, Manrubia S (2014) Large-scale genomic analysis suggests a neutral punctuated dynamics of transposable elements in bacterial genomes. PLoS Comput Biol 10:1–11
Jacobson JW, Medhora MM, Hartl DL (1986) Molecular structure of a somatically unstable transposable element in Drosophila. Proc Natl Acad Sci U S A 83:8684–8688
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian Protein Metabolism. Academic Press, New York, pp 21–132
Krieger MJB, Ross KG (2003) Molecular evolutionary analyses of mariners and other transposable elements in fire ants (Hymenoptera, Formicidae). Insect Mol Biol 12:155–165
Lahr DJG, Katz LA (2009) Reducing the impact of PCR-mediated recombination in molecular evolution and environmental studies using a new-generation high-fidelity DNA polymerase. Biotechniques 47:857–866
Lampe DJ, Witherspoon DJ, Soto-Adames FN, Robertson HM (2003) Recent horizontal transfer of mellifera subfamily mariner transposons into insect lineages representing four different orders shows that selection acts only during horizontal transfer. Mol Biol Evol 20:554–562
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Loreto EL, Carareto CM, Capy P (2008) Revisiting horizontal transfer of transposable elements in Drosophila. Heredity 100:545–554
Lorite P, Carrillo JA, Garneria I, Petitpierre E, Palomeque T (2002) Satellite DNA in the elm leaf beetle Xanthogaleruca luteola (Coleoptera, Chrysomelidae): characterization, interpopulation analysis and chromosome location. Cytogenet Genome Res 98:302–307
Lorite P, Maside X, Sanllorente O, Torres MI, Periquet G, Palomeque T (2012) The ant genomes have been invaded several times by mariner transposable elements. Naturwissenschaften 12:1007–1020
Lu C, Chen J, Zhang Y, Hu Q, Su W, Kuang H (2012) Miniature inverted–repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol Biol Evol 29:1005–1017
Marzo M, Liu D, Ruiz A, Chalmers R (2013) Identification of multiple binding sites for the THAP domain of the Galileo transposase in the long terminal inverted-repeats. Gene 525:84–91
Moreau CS, Bell CD, Vila R, Archibald SB, Pierce NE (2006) Phylogeny of the ants: Diversification in the age of Angiosperms. Science 312:101–104
Muñoz-López M, Siddique A, Bischerour J, Lorite P, Chalmers R, Palomeque T (2008) Transposition of Mboumar-9: identification of a new naturally-active mariner-family transposón. J Mol Biol 382:567–572
Muñoz-Torres MC, Reese JT, Childers CP, Bennett AK, Sundaram JP, Childs KL, Anzola JM, Milshina N, Elsik CG (2011) Hymenoptera Genome Database: integrated community resources for insect species of the order Hymenoptera. Nucleic Acids Res 39 (suppl 1):D658–D662
Nei M (1987) Molecular Evolutionary Genetics. Columbia Univ. Press, New York
Nygaard S, Wurm Y (2015) Ant genomics (Hymenoptera: Formicidae): challenges to overcome and opportunities to seize. Myrmecological News 21:59–72
Nygaard S, Zhang G, Schiøtt M, Li C, Wurm Y, Hu H, Zhou J, Ji L, Qiu F, Rasmussen M, Pan H, Hauser F, Krogh A, Grimmelikhuijzen CJ, Wang J, Boomsma JJ (2011) The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming. Genome Res 21:1339–1348
Ouellette GD, Fisher BL, Girman DJ (2006) Molecular systematics of basal subfamilies of ants using 28S rRNA (Hymenoptera: Formicidae). Mol Phylogenet Evol 40:359–369
Oxley PR, Ji L, Fetter-Pruneda I, McKenzie SK, Li C, Hu H, Zhang G, Kronauer DJ (2014) The genome of the clonal raider ant Cerapachys biroi. Curr Biol 24:451–458
Palomeque T, Muñoz-López M, Carrillo JA, Lorite P (2005) Characterization and evolutionary dynamics of a complex family of satellite DNA in the leaf beetle Chrysolina carnifex (Coleoptera, Chrysomelidae). Chromosom Res 13:795–807
Palomeque T, Carrillo JA, Muñoz-López M, Lorite P (2006) Detection of a mariner-like element and a miniature inverted-repeat transposable element (MITE) associated with the heterochromatin from ants of the genus Messor and their possible involvement for satellite DNA evolution. Gene 371:194–205
Perrichot V, Lacau S, Néraudeau D, Nel A (2008) Fossil evidence for the early ant evolution. Naturwissenschaften 95:85–90
Robertson HM (2002) Evolution of DNA transposons in eukaryotes. In: Craig NL, Craigie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. American Society for Microbiology Press, Washington DC, pp 1093–1110
Robertson HM, Asplund KL (1996) Bmmar1, a basal lineage of the mariner family of transposable elements in the silkworm moth, Bombyx mori. Insect Biochem Mol Biol 26:945–954
Rouleux-Bonnin F, Petit A, Demattei MV, Bigot Y (2005) Evolution of full-length and deleted forms of the mariner-like element, Botmar1, in the genome of the bumble bee, Bombus terrestris (Hymenoptera: Apidae). J Mol Evol 60:736–747
Schaack S, Gilbert C, Feschotte C (2010) Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol 25:537–546
Serra F, Becher V, Dopazo H (2013) Neutral theory predicts the relative abundance and diversity of genetic elements in a broad array of eukaryotic genomes. PLoS One 8:1–9
Shimodaira H (2002) An approximately unbiased test of phylogenetic tree selection. Syst Biol 51:492–508
Shimodaira H, Hasegawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol 16:1114–1116
Simola DF, Wissler L, Donahue G, Waterhouse RM, Helmkampf M, Roux J, Nygaard S, Glastad KM, Hagen DE, Viljakainen L, Reese JT, Hunt BG, Graur D, Elhaik E, Kriventseva EV, Wen J, Parker BJ, Cash E, Privman E, Childers CP, Muñoz-Torres MC, Boomsma JJ, Bornberg-Bauer E, Currie CR, Elsik CG, Suen G, Goodisman MA, Keller L, Liebig J, Rawls A, Reinberg D, Smith CD, Smith CR, Tsutsui N, Wurm Y, Zdobnov EM, Berger SL, Gadau J (2013) Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. Genome Res 23:1235–1247
Smith CD, Zimin A, Holt C, Abouheif E, Benton R, Cash E, Croset V, Currie CR, Elhaik E, Elsik CG, Fave MJ, Fernandes V, Gadau J, Gibson JD, Graur D, Grubbs KJ, Hagen DE, Helmkampf M, Holley JA, Hu H, Viniegra AS, Johnson BR, Johnson RM, Khila A, Kim JW, Laird J, Mathis KA, Moeller JA, Muñoz-Torres MC, Murphy MC, Nakamura R, Nigam S, Overson RP, Placek JE, Rajakumar R, Reese JT, Robertson HM, Smith CR, Suarez AV, Suen G, Suhr EL, Tao S, Torres CW, van Wilgenburg E, Viljakainen L, Walden KK, Wild AL, Yandell M, Yorke JA, Tsutsui ND (2011a) Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc Natl Acad Sci U S A 108:5673–5678
Smith CR, Smith CD, Robertson HM, Helmkampf M, Zimin A, Yandell M, Holt C, Hu H, Abouheif E, Benton R, Cash E, Croset V, Currie CR, Elhaik E, Elsik CG, Favé MJ, Fernandes V, Gibson JD, Graur D, Gronenberg W, Grubbs KJ, Hagen DE, Viniegra AS, Johnson BR, Johnson RM, Khila A, Kim JW, Mathis KA, Munoz-Torres MC, Murphy MC, Mustard JA, Nakamura R, Niehuis O, Nigam S, Overson RP, Placek JE, Rajakumar R, Reese JT, Suen G, Tao S, Torres CW, Tsutsui ND, Viljakainen L, Wolschin F, Gadau J (2011b) Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc Natl Acad Sci U S A 108:5667–5672
Suen G, Teiling C, Li L, Holt C, Abouheif E, Bornberg-Bauer E, Bouffard P, Caldera EJ, Cash E, Cavanaugh A, Denas O, Elhaik E, Favé MJ, Gadau J, Gibson JD, Graur D, Grubbs KJ, Hagen DE, Harkins TT, Helmkampf M, Hu H, Johnson BR, Kim J, Marsh SE, Moeller JA, Muñoz-Torres MC, Murphy MC, Naughton MC, Nigam S, Overson R, Rajakumar R, Reese JT, Scott JJ, Smith CR, Tao S, Tsutsui ND, Viljakainen L, Wissler L, Yandell MD, Zimmer F, Taylor J, Slater SC, Clifton SW, Warren WC, Elsik CG, Smith CD, Weinstock GM, Gerardo NM, Currie CR (2011) The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLoS Genet 7:e1002007
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Valles SM, Oi DH, Yu F, Tan XX, Buss EA (2012) Metatranscriptomics and pyrosequencing facilitate discovery of potential viral natural enemies of the invasive Caribbean crazy ant, Nylanderia pubens. PLoS One 7:e31828
Varriale A (2014) DNA methylation, epigenetics, and evolution in vertebrates: facts and challenges. Int J Evol Biol 2014: ID 475981
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276
Wurm Y, Uva P, Ricci F, Wang J, Jemielity S, Iseli C, Falquet L, Keller L (2009) Fourmidable: a database for ant genomics. BMC Genomics 10:5
Wurm Y, Wang J, Riba-Grognuz O, Corona M, Nygaard S, Hunt BG, Ingram KK, Falquet L, Nipitwattanaphon M, Gotzek D, Dijkstra MB, Oettler J, Comtesse F, Shih CJ, Wu WJ, Yang CC, Thomas J, Beaudoing E, Pradervand S, Flegel V, Cook ED, Fabbretti R, Stockinger H, Long L, Farmerie WG, Oakey J, Boomsma JJ, Pamilo P, Yi SV, Heinze J, Goodisman MA, Farinelli L, Harshman K, Hulo N, Cerutti L, Xenarios I, Shoemaker D, Keller L (2011) The genome of the fire ant Solenopsis invicta. Proc Natl Acad Sci U S A 108:5679–5684
Yang G, Nagel DH, Feschotte C, Hankock CN, Wesslerr SR (2009) Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science 325:1391–1394
Acknowledgments
This work was supported by the Spanish Junta de Andalucía (through the programs “Ayudas a Grupos de Investigación,” Group BIO220 and “Incentivos a proyectos de investigación de excelencia,” project CVI-6807, co-funded by the European Regional Development Fund) and by the Spanish Ministerio de Educación e Innovación (through project CGL2011-23841, co-funded by the European Regional Development Fund).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Sven Thatje
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 1430 kb)
Rights and permissions
About this article
Cite this article
Palomeque, T., Sanllorente, O., Maside, X. et al. Evolutionary history of the Azteca-like mariner transposons and their host ants. Sci Nat 102, 44 (2015). https://doi.org/10.1007/s00114-015-1294-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00114-015-1294-3