Abstract
Joint homeostasis failure can result in osteoarthritis (OA). Currently, there are no treatments to alter disease progression in OA, but targeting early changes in cellular behavior has great potential. Recent data show that nuclear receptors contribute to the pathogenesis of OA and could be viable therapeutic targets, but their molecular mechanisms in cartilage are incompletely understood. This study examines global changes in gene expression after treatment with agonists for four nuclear receptor implicated in OA (LXR, PPARδ, PPARγ, and RXR). Murine articular chondrocytes were treated with agonists for LXR, PPARδ, PPARγ, or RXR and underwent microarray, qPCR, and cellular lipid analyses to evaluate changes in gene expression and lipid profile. Immunohistochemistry was conducted to compare two differentially expressed targets (Txnip, Gsta4) in control and cartilage-specific PPARδ knockout mice subjected to surgical destabilization of the medial meniscus (DMM). Nuclear receptor agonists induced different gene expression profiles with many responses affecting lipid metabolism. LXR activation downregulated gene expression of proteases involved in OA, whereas RXR agonism decreased expression of ECM components and increased expression of Mmp13. Functional assays indicate increases in cell triglyceride accumulation after PPARγ, LXR, and RXR agonism but a decrease after PPARδ agonism. PPARδ and RXR downregulate the antioxidant Gsta4, and PPARδ upregulates Txnip. Wild-type, but not PPARδ-deficient mice, display increased staining for Txnip after DMM. Collectively, these data demonstrate that nuclear receptor activation in chondrocytes primarily affects lipid metabolism. In the case of PPARδ, this change might lead to increased oxidative stress, possibly contributing to OA-associated changes.
Key message
-
Nuclear receptors regulate metabolic genes in chondrocytes.
-
Nuclear receptors affect triglyceride levels.
-
PPARδ mediates regulation of oxidative stress markers.
-
Nuclear receptors are promising therapeutic targets for osteoarthritis.
Similar content being viewed by others
Introduction
Dysregulation of joint homeostasis can result in osteoarthritis (OA), a collective of heterogeneous pathologies culminating in joint failure. OA presents with similar pathological end points, but mechanisms of initiation and progression vary among subtypes of this disease, which is one of the leading causes of disability worldwide [1, 2]. Its varied presentation influences whether it is symptomatic or not and even whether it can be diagnosed radiographically. Multiple tissues, such as the articular cartilage, subchondral bone, synovium, meniscus, and fat pads, are involved in this condition, and initiation of this disease can stem from mechanical, metabolic, or age-associated factors, although none of these are mutually exclusive.
The main function of the cartilage is to act as a shock absorber, mediating load bearing through the influx and efflux of water attracted to the proteoglycan aggregates of the extracellular matrix and through the tensile strength conferred by collagen fibril organization [3]. Although cartilage cells contribute a small percentage of the volume of the entire joint, they are sensitive to external factors and respond with changes in gene expression affecting OA, thus underscoring their importance in joint homeostasis.
Metabolic OA has been classified as a distinct subtype of OA associated with disorders such as dyslipidemia, hypertension, and obesity [4]. Imbalances in systemic lipid and cholesterol metabolism, nutrient exchange, accumulation of advanced glycation end products, and increases in adipokines contribute to this condition. Changes in lipid metabolism, in particular, may directly affect joint homeostasis through ectopic lipid deposition in chondrocytes [4–6]. In fact, both chondrocyte-specific cholesterol accumulation and high-fat diet have caused increased disease severity in murine models [7–9]. Altogether, these data suggest direct regulation of cartilage homeostasis by lipid metabolism.
Nuclear receptors are a class of proteins that are activated by small molecule ligands and can up- or downregulate the expression of target genes through the recruitment of co-factors. They have been reported as attractive potential targets for pharmacological therapy because of their ability to bind synthetic or natural ligands that regulate transcriptional activity [10]. As such, synthetic agonists for nuclear receptors have been developed to target metabolic conditions such as dyslipidemia, atherosclerosis, and diabetes [11, 12]. Peroxisome proliferated activated receptors (PPARs) are typically involved in the control of lipid metabolism and activated by the binding of endogenous fatty acids, whereas liver X receptor (LXR) is principally involved in cholesterol metabolism. Recently, we have shown that cartilage-specific ablation of the gene encoding the nuclear receptor PPARδ has a protective effect on cartilage after surgical induction of OA, demonstrating that PPARδ promotes post-traumatic OA. Conversely, PPARγ and LXR are protective and necessary for normal joint function and skeletal development [13–16]. Interestingly, all three of these receptors act in heterodimers with the common partner RXR, positioning RXR at the center of a complex network of nuclear receptors. All of these proteins are expressed in the cartilage [15, 17, 18]. However, the mode of action of these proteins in the cartilage is largely unknown, and since they are transcription factors, identification of their target genes is essential to understand their specific roles and to evaluate their value as therapeutic targets. Here, we attempted to identify these target genes in a genome-wide manner.
In this study, we have used microarray analysis paired with functional validation to identify gene targets of LXR, PPARγ, PPARδ, and RXR in articular chondrocytes, in order to elucidate their potential role in OA pathogenesis. There is strong evidence implicating the involvement of nuclear receptors in the progression or prevention of OA, and here, we provide insight as to how they may be involved in altering the gene expression profile and phenotype of mature, healthy chondrocyte cultures. We are also the first, to our knowledge, to quantify changes in neutral lipid and free cholesterol mass in chondrocytes in vitro. This information is essential in uncovering the early changes that occur in chondrocytes before irreversible phenotypic changes within the joint and is vital since we currently have no effective biomarkers or treatment to alter the course of OA progression. Our data demonstrate that changes in gene regulation after nuclear receptor agonist treatment primarily affect lipid metabolism, suggesting a close link between lipid metabolism within chondrocytes and the progression of OA.
Methods
Primary cell culture and isolation
Immature murine articular chondrocytes (IMACs) were isolated from the femoral head, femoral condyle, and tibial condyles of 5–6-day-old CD1 mice (Charles River Laboratories) as per [19]. The tissue was then subjected to 1 h (3 mg/ml) followed by 24 h (0.5 mg/ml) incubations in Collagenase D diluted in Dulbecco’s Modified Eagles Medium supplemented with 2 mM l-glutamine, 50 U/ml penicillin, and 0.05 mg/ml streptomycin at 37 °C under 5% CO2. The tissue fragments were then agitated, and cells were isolated and cultured [19]. On the seventh day after isolation, cells were treated with either PPARδ agonist (GW501516), PPARγ agonist (Rosiglitazone), LXR agonist (GW3965), RXR agonist (SR11237), or control (DMSO), all at concentrations of 1 μM for 72 h. Pyruvate dehydrogenase kinase (PDK) inhibition studies were done using these nuclear agonist treatments and one of two inhibitors at 5 μM: dichloroacetate (DCA, pan-PDK inhibitor) or diisopropylaminedichloroacetate (DADA, PDK4 inhibitor), or vehicle control (water).
RNA extraction, purification, and qPCR
Total RNA was isolated from cells using TRIzol® (Invitrogen). Cells were lysed in TRIzol® reagent, phases were separated using chloroform (20%), and supernatant was removed. RNA was precipitated using 100% isopropanol (0.5%) and washed using 70% ethanol followed by air-drying and resuspension in RNAse free water, as per manufacturer’s instructions. RNA was quantified using a Nanodrop 2000 spectrophotometer. RNA integrity was confirmed with Aligent 2100 BioAnalyzer Data Review Software (Wilmington, DE) at the London Regional Genomics Centre. Samples with RNA integrity number (RIN) values greater than 8 were used for microarray analysis.
Real-time PCR (qPCR) was performed as per [16]. In brief, qPCR was performed using a One-Step RT qPCR Master Mix kit and TaqMan Gene Expression Assays (Applied Biosystems), with 40 cycles on an ABI Prism 7900HT sequence detector (PerkinElmer), or on a Bio-Rad CFX384 RT-PCR system with 10–15 μl reaction volumes of iQ SYBR Green Supermix (Biorad) with diluted cDNA equivalent to 200–500 ng of input RNA per reaction, as well as 25–50 μM forward and reverse primers [20]. Probes for Acan(Mm00545794_m1), Actb (Mm02619580_g1), Adamts4(Mm00556068_m1), Adamts5 (Mm00478620_m1), Angptl4(Mm00480431_m1), Col2a1 (Mm01309565_m1), Fabp3(Mm02342495_m1), Fabp4(Mm00445878_m1), LPL(Mm00434764_m1), Mmp2 (Mm00439498_m1), Mmp3 (Mm00440295_m1), Mmp13(Mm00439491_m1), Pdk4(Mm01166879_m1), and Sox9 (Mm00448840_m1) were purchased from Life Technologies. Gene expression was normalized relative to Actb or 18S (viability studies only). Relative gene expression was calculated using the ΔΔCt method [21] as described [22]. Statistical analysis was conducted using GraphPad Prism 6.0. Values were transformed, and a one-way analysis of variance (ANOVA) was performed followed by Tukey’s multiple comparisons tests.
Microarray and data analysis
Total RNA (200 ng per sample) was subject to 2 rounds of amplification followed by labeling and hybridization to Affymetrix GeneChip® Mouse Gene 2.0 ST Array containing 35,240 probes at the London Regional Genomics Centre (London, Ontario, Canada) as described [23]. Three independent cell and RNA isolations were used for each treatment. Probe data was analyzed, and gene level, ANOVA p values, and fold changes were determined using Partek Genomics Suite v6.6. Genes with at least 1.5-fold change, with p < 0.05 were considered significant and used for subsequent analyses. The complete array data set will be publicly available through Gene Expression Omnibus (GEO). The Venn diagrams were created using the online plotting tool Venny 2.0.1 [24]. KEGG pathway maps were generated using Ingenuity Pathway Analysis. Gene ontology biological processes and cellular component processes were classified through GO consortium available at geneontology.org using the PANTHER Overrepresentation Test (released 2016-07-15) and GO Ontology Database (released 2016-10-27), Mus musculus reference list, and Bonferroni correction for multiple testing. Biological processes identified with more than three genes involved were included in the table.
Cellular lipid mass
IMACs were isolated, cultured, and treated with nuclear receptor agonists as described above. At the 72 h time point, cells were washed with 0.2% BSA in phosphate-buffered saline (PBS), followed by three washes in PBS. Lipids were extracted using 3:2 hexane/isopropanol solvent and pooled. Hexane/isopropanol solvents were evaporated to dryness under N2 and resuspended in 1.4 ml of chloroform–triton (0.5% triton v/v). Solvent was re-evaporated, and lipids were re-solubilized in 350 μl water. Two 50-μl aliquots were used per sample to determine total cholesterol (TC), free cholesterol (FC), and triglyceride (TG) mass, spectrophotometrically as per [25]. Cholesteryl esters (CE) were calculated by subtracting FC from TC. Proteins were extracted using 0.2 NaOH overnight incubation to digest chondrocytes and quantified using a standard BCA protein assay (Pierce, Thermo Fisher Scientific). All cell lipid measures reported are standardized to milligrams of cell protein. Values were normalized relative to vehicle control DMSO, and statistical analyses were performed using GraphPad Prism 6.0. Values were transformed, and a one-way analysis of variance (ANOVA) was performed followed by Tukey’s multiple comparisons tests.
Animals and surgery
All animal experiments were approved by the Animal Use Subcommittee at The University of Western Ontario and were conducted in accordance with the guidelines from the Canadian Council on Animal Care. Mice were group housed (6 mice per cage) in colony cages on a standard 12 h light/dark cycle with free access to standard mouse chow, water, and running wheels. Surgical destabilization of the medial meniscus (DMM) or SHAM surgery was performed on 12-week-old C57BL/6 male mice (N = 8–9 per group), as described in [13]. Mice were euthanized at 10 or 12 weeks post-surgery for preparation of paraffin sections and subsequent histological analysis. Another cohort of 20-week-old male cartilage-specific Ppard knockout mice and wild-type littermate controls underwent DMM surgery (N = 5 per group) and was harvested for histological analyses 8 weeks later as in [13]. Ppard mutant mice were bred and genotypes as described in [13]. Paraffin sections from these studies were employed to evaluate the presence of Thioredoxin Interacting Protein (Txnip) and glutathione S-transferase A4 (Gsta4).
Immunohistochemistry
Immunohistochemistry was performed on frontal sections of paraffin-embedded knee joints as described [26]. Txnip rabbit polyclonal antibody was purchased from Proteintech (18243-1-AP). Slides without primary antibody were used as controls, antigen retrieval was performed in 0.1% Triton in H20, and primary antibody was used at a concentration of 1:300. Gsta4 rabbit polyclonal antibody was purchased from Proteintech (17271-1-AP), and immunohistochemical staining was performed under the same conditions as above, except with a primary antibody concentration of 1:100.
Results
Global changes in chondrocyte gene expression in response to nuclear receptor agonists
We have previously reported that treatment of articular chondrocytes with the PPARδ agonist GW501516 results in increased catabolic gene expression and robust fatty acid oxidation. We have also determined that the LXR agonist GW3965 delays chondrocyte hypertrophy [13, 16]. Identifying which genes are responsible for these phenotypes and how they interact with each other is key to understanding signaling pathways responsible for joint homeostasis and the prevention of osteoarthritis. We first examined global changes in chondrocyte gene expression in response to 1 μM treatment with LXR agonist GW3965, RXR agonist SR11237, PPARδ agonist GW501516, or PPARγ agonist Rosiglitazone. RNA was isolated from IMACs cultured with agonists for 72 h, then hybridized to Affymetrix microarrays representing the mouse genome.
We compiled a list of genes changed by more than 1.5-fold (refer to supplementary data for full list). LXR agonism significantly altered 128 genes (97 upregulated, 31 downregulated), RXR agonism differentially regulated a total of 108 genes (67 upregulated, 41 downregulated), PPARδ agonism induced changes in 58 genes (48 upregulated, 10 downregulated), while PPARγ agonism changed 32 genes (29 upregulated, 3 downregulated). The most robust and significantly upregulated and downregulated genes after nuclear receptor agonist treatment are shown in Fig. 1.
Microarray analyses of nuclear receptor agonist effects on chondrocyte gene expression. Microarray analysis of RNA isolated from immature murine articular chondrocytes treated for 72 h with 1 μM LXR agonist GW3965 (a), RXR agonist SR11237 (b), PPARδ agonist GW501516 (c), or PPARγ agonist Rosiglitazone (d). The most highly upregulated and downregulated genes are shown with fold change relative to vehicle control DMSO (1 μM)
Nuclear receptors affect common biological functions in chondrocytes
Comparison of gene expression profiles induced by various nuclear receptor agonists revealed several common hits. We therefore decided to evaluate shared functional roles by identifying similar biological processes through Gene Ontology. Supplementary Table 1 indicates the biological processes regulated by agonist treatment for each nuclear receptor, and common processes are highlighted with the same color. Both LXR and RXR agonism altered cholesterol biosynthetic processes, while LXR and PPARγ regulated triglyceride metabolism, and RXR and PPARγ increased metabolic processes in chondrocytes. We also investigated GO cellular component processes affected by each agonist (Supplementary Table 2). We noted that the extracellular matrix, mitochondrion, mitochondrial membrane, and endoplasmic reticulum were key areas in these processes, and that lipid particles and extracellular space were common between at least two treatment groups. The identified cellular components were consistent with our biological processes, heavily implicating the metabolic functions of these nuclear receptor agonists. In order to compare relationships between nuclear receptor agonist treatments, we created a Venn diagram to illustrate the number of genes induced by multiple receptors (Fig. 2a). The two genes upregulated by all four nuclear receptor agonists were Pdk4 and Angptl4. Pdk4 functions as an inhibitor of the pyruvate dehydrogenase complex. Thus, it plays a key regulatory role in shifting energy utilization from glycolytic to fatty acid metabolism in the cell [27]. Angptl4 is a well-known direct target of PPARs that is upregulated by hypoxia and has been characterized as an adipocytokine [28]. It has also been identified as a potential pro-angiogenic mediator of arthritis, is involved in ECM remodeling, and is upregulated in the cartilage of RA and OA patients [29–31]. Larger numbers of genes were regulated by two or three different agonists (Fig. 2a), and differences in gene regulation between the commonly induced genes are hierarchically clustered and visually represented in a heatmap (Fig. 2b).
Comparison of nuclear receptor agonist effects on chondrocyte gene expression. a Comparison of all genes regulated by the four different nuclear receptor agonists on chondrocytes demonstrates that two genes are commonly regulated by all four nuclear receptors. Nine genes are commonly regulated by LXR, PPARδ, and RXR, while four genes are commonly regulated by LXR, PPARδ, and PPAR γ. Three genes are regulated by LXR, PPARγ, and RXR, and two genes are commonly regulated by PPARδ, PPARγ, and RXR. b Differences in regulation of genes commonly changed by all four nuclear receptor agonism are analyzed through hierarchical clustering and visually displayed in a heatmap
KEGG pathway analysis was subsequently conducted to identify whether the genes common among agonist treatment would be associated with shared processes. We discovered that the four agonists shared 13 common pathways (Fig. 3). Of these, the most significantly enriched pathways included the PPAR signaling pathway and the adipocytokine signaling pathway (Supplementary Figs. 1 and 2). According to our analyses, all four nuclear receptors are involved in adipocyte differentiation and fatty acid transport. While LXR, PPARγ, and RXR regulate lipogenesis and LXR, PPARδ, and RXR mediate fatty acid oxidation and beta oxidation, only PPARδ agonism induces the ketogenic pathway.
Comparison of nuclear receptor agonist effects on affected KEGG pathways. Comparison of all KEGG pathways enriched by the four different nuclear receptor agonist treatments in chondrocytes demonstrates that 13 pathways are commonly enriched by all four nuclear receptors. Seventeen pathways are commonly enriched by LXR, PPARδ, and RXR, while two pathways are commonly enriched by LXR, PPARδ, and PPARγ. Five pathways are commonly enriched by LXR, PPARγ, and RXR, and no pathways are commonly enriched by PPARδ, PPARγ, and RXR
Since PPARδ has opposite effects on OA progression than PPARγ and LXR, we were particularly interested in genes showing opposite responses to the respective agonists. However, the only gene that was differentially regulated (at our selected threshold) between any of the treatments was Txnip, which encodes the Thioredoxin interacting protein. Txnip inhibits Thioredoxin and contributes to ER stress, inflammasome activation, and the accumulation of reactive oxygen species (ROS) [32]. This gene was upregulated by PPARδ agonist GW501516 and downregulated by LXR agonist GW3965 treatment (Fig. 2b). Based on the common pathways and genes identified, we next validated changes in the expression of selected genes by qPCR.
LXR, RXR, and PPAR agonism promote changes in genes involved in ECM homeostasis and chondrocyte metabolism
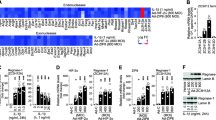
Genes induced in the microarray were primarily involved in metabolic processes or in extracellular matrix component production and turnover. We chose to validate a subset of these genes that were shared between nuclear receptor agonist treatments. Aggrecan and Fibrillin 2 are extracellular matrix proteins encoded by the Acan and Fbn2 genes. In concert with our microarray results, gene expression of Acan was significantly lower than vehicle control (DMSO) with RXR agonist treatment. Similarly, both LXR and RXR agonism significantly lowered gene expression of Fbn2 (Fig. 4). Gene expression of Collagen 2 (Col2a1) remained unchanged in response to any of the treatments. Next, we validated expression of protease genes that were changed by some of the nuclear receptor agonists and accordingly found that gene expression of Adamts4, Mmp2, and Mmp13 were significantly reduced by LXR agonism. Interestingly, RXR agonism decreased gene expression of Adamts4 while increasing that of Mmp13 (the primary collagenase of OA), implying a selective pathway for ECM remodeling and degradation.
Effects of nuclear receptor agonist treatments on extracellular matrix gene expression in chondrocytes. IMACs were incubated for 72 h with 1 μM DMSO (vehicle control), PPARδ agonist GW501516, PPARγ agonist Rosiglitazone, LXR agonist GW3965, or RXR agonist SR11237. a Relative gene expression of Acan gene is significantly reduced by treatment with the RXR agonist. b, c Relative gene expression of Adamts4 and Fbn2 are significantly reduced by LXR and RXR agonist treatment. d, e Relative gene expression of matrix metalloproteinases Mmp2 and Mmp13 is decreased by LXR agonist treatment, while gene expression of Mmp13 is significantly elevated by RXR agonist treatment. f Col2a1 gene expression remains unchanged by all treatments. Values represented are the mean ± SEM of ≥3 independent cell isolations. *p < 0.05
LXR, RXR, and PPARs are involved in the regulation of metabolism in a number of tissues. In a previous study, we showed that chondrocytes express functional PPARδ and are capable of responding to GW501516 stimulation with increased fatty acid oxidation [13]. All four nuclear receptor agonists induced strong effects on genes encoding metabolic enzymes. Angptl4 and Pdk4, the two common genes induced by all four nuclear receptors in the microarray, demonstrated a similar robust upregulation in qPCR validation (Fig. 5). Abca1, Cidea, Cpt1a, Lpl, and Insig1 were significantly increased by PPARδ, LXR, and RXR agonist treatment, and LXR also significantly increased the expression of Srebf1. Gene expression of cytoskeletal fatty acid transporter Fabp3 was significantly increased by PPARδ activation, while Gsta4, a gene encoding an enzyme for cellular defense against reactive electrophiles [33], was significantly reduced by both PPARδ and RXR agonism.
Effects of nuclear receptor agonist treatment on metabolic gene expression in chondrocytes. IMACs were incubated for 72 h with 1 μM DMSO (vehicle control), PPARδ agonist GW501516, PPARγ agonist Rosiglitazone, LXR agonist GW3965, or RXR agonist SR11237. a, c, d, g, i Relative gene expression of Abca1, Cidea, Cpt1a, Lpl, and Insig1 is significantly increased by PPARδ, LXR, and RXR treatments. b, f Relative gene expression of Angptl4 and Pdk4 is elevated by all four treatments. e Relative gene expression of Fabp3 is significantly upregulated by PPARδ agonism only. g RXR and PPARδ treatment significantly decreased relative gene expression of Gsta4. h Srebf1 expression is significantly upregulated by LXR agonism only. Values represented are the mean ± SEM of ≥3 independent cell isolations. *p < 0.05
In a previous study, our group has examined the toxicity of the administration of PPARδ agonist GW501516; we have shown that it does not alter cell number [13], while LXR stimulation increases cell number [16]. Here, we examined whether administration of any of the four nuclear receptor agonists affects cell physiology through changes in gene expression of markers for proliferation (Ccnd1c, Pcna), cell cycle exit (p57/Cdkn1c), apoptosis (Bax), or hypertrophic differentiation (ColX, Runx2) (Supplementary Fig. 3). We do not see any changes in these parameters, with the exception of RXR agonism decreasing the expression of Runx2.
Increased expression of oxidative stress markers in osteoarthritic cartilage
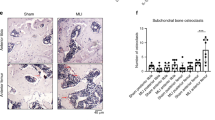
Txnip plays an important regulatory role in mediating oxidative stress and inflammation in a number of tissues [32]. Txnip was the only gene differentially regulated between nuclear receptor agonists. LXR agonist treatment downregulated gene expression, while PPARδ highly induced Txnip. These patterns observed in microarray analyses were evaluated by qPCR, where PPARδ agonism significantly increased gene expression of Txnip, while LXR agonist-treated cells demonstrated trends toward decreased gene expression, and RXR and PPARγ agonism showed no change (Fig. 6a). To examine whether Txnip expression is linked to OA, immunohistochemistry for Txnip was performed on frontal sections of mice after DMM surgery (Fig. 6b, c). Wild-type mice 10 weeks post-surgery had increased staining in remaining cartilage compared to mice that underwent sham surgery. To validate the effects of PPARδ on Txnip expression, we compared protein expression in cartilage-specific Ppard KO mice and wild-type littermates 8 weeks after DMM surgery. Wild-type mice demonstrated increased staining for Txnip after DMM surgery, particularly in areas of osteophyte growth at joint margins, whereas both sham-operated control mice and KO mice after either surgery showed little to no staining. The apparent increase of Txnip expression in the process of OA implies an imbalance in regulatory processes governing oxidative stress and inflammation, potentially linking changes in metabolism to osteoarthritic changes.
Effects of nuclear receptor agonist treatment on Txnip expression. a IMACs were incubated for 72 h with 1 μM DMSO (vehicle control), PPARδ agonist GW501516, PPARγ agonist Rosiglitazone, LXR agonist GW3965, or RXR agonist SR11237. PPARδ treatment significantly increased gene expression of Txnip. Values represented are the mean ± SEM of 4 independent cell isolations. *p < 0.05. b Immunohistochemistry for Txnip demonstrates increased cellular staining in the cartilage of WT DMM mice 10 weeks post-surgery relative to sham mice. c Immunohistochemical staining for Txnip in cartilage-specific Ppard KO mice vs WT littermate controls 8 weeks post DMM surgery. Ppard KO mice display less staining than WT mice
In our qPCR validation of target genes, we saw decreased expression of Gsta4 (Fig. 7a) after PPARδ and RXR agonist treatment. Gsta4 protects the cell from reactive aldehydes that are produced as a result of lipid peroxidation or oxidative stress. We assessed the localization of Gsta4 in the DMM model and found that sham-operated animals displayed consistent immunohistochemical staining in superficial cartilage and meniscus, both in wild-type and cartilage-specific Ppard KO mice (Fig. 7b, c). DMM-operated animals showed little to no staining in cartilage of both genotypes, even if superficial cartilage remained intact.
Effects of nuclear receptor agonist treatment on Gsta4 expression. a IMACs were incubated for 72 h with 1 μM DMSO (vehicle control), PPARδ agonist GW501516, PPARγ agonist Rosiglitazone, LXR agonist GW3965, or RXR agonist SR11237. PPARδ agonist treatment and RXR agonist treatment both significantly decreased gene expression of Gsta4. Values represented are the mean ± SEM of 4 independent cell isolations. *p < 0.05. b Immunohistochemistry for Gsta4 demonstrates decreased cellular staining in the cartilage of WT DMM mice 12 weeks post-surgery relative to sham mice. c Immunohistochemical staining for Gsta4 in cartilage-specific Ppard KO mice vs WT littermate controls 8 weeks post-DMM or sham surgery. Both WT and Ppard KO mice display little to no staining of Gsta4 in the articular cartilage after DMM surgery, compared to sham controls.
Changes in gene expression correspond with functional changes in chondrocyte lipid profile
In light of the number of genes involved in lipid metabolism that were identified in our gene expression analyses, we assessed neutral lipid and cholesterol accumulation in chondrocytes. Using the same nuclear receptor agonist treatment protocols, we harvested IMACs for cellular lipid mass assays. These assays allowed us to directly quantify triglycerides and cholesterol in vitro. There were significant changes in cell triglycerides, but not total cholesterol, free cholesterol, or cholesteryl esters (Fig. 8). These data suggest that changes in lipid metabolism upon agonist treatment are likely related to lipogenesis and fatty acid oxidation, rather than cholesterol transport or accumulation. In particular, triglycerides were significantly decreased with PPARδ agonist treatment and were significantly elevated with LXR, PPARγ, and RXR agonism. These changes are consistent with the known effects of activation of these nuclear receptors on triglyceride metabolism in other cell types and suggest that PPARδ may have an opposing role in lipid metabolism in chondrocytes relative to the other nuclear receptors examined [34].
Quantification of cellular lipid mass in nuclear receptor agonist-treated chondrocytes. IMACs were incubated for 72 h with 1 μM DMSO (vehicle control), PPARδ agonist GW501516, PPARγ agonist Rosiglitazone, LXR agonist GW3965, or RXR agonist SR11237. Lipids were extracted, isolated, and mass was measured spectrophotometrically. Proteins were isolated and quantified using BCA. Measurements are reported relative to mg of cell protein. Absolute mean values for triglycerides, total cholesterol, free cholesterol, and cholesterol esters were 7.4, 27.1, 26.4, and 0.75 μg/mg protein, respectively, for DMSO-treated cells. a Cell triglycerides (μg) are significantly elevated by PPARγ, LXR, and RXR treatment and are significantly decreased by PPARδ agonism. b, c, d Total cholesterol, free cholesterol, and cholesterol ester remain unchanged after nuclear receptor agonist treatment. Values represented are the mean ± SEM of ≥5 independent cell isolations. *p < 0.05
We investigated whether altering metabolic pathways would affect lipid metabolism in chondrocytes treated with the nuclear receptor agonists. Given the effects of all agonists on Pdk4, we performed inhibition of pyruvate dehydrogenase kinase with a pan-PDK inhibitor (DCA) or PDK4 specific inhibitor (DADA) (Supplementary Fig. 4). PDK enzymes act to inhibit pyruvate dehydrogenase which catalyzes the first step of the pyruvate dehydrogenase complex (PDC). The PDC oxidizes pyruvate to generate acetyl-coA to be used in the TCA cycle, thereby promoting the preferential oxidation of glucose. Either treatment did not affect the responses of chondrocyte triglyceride levels to the four agonists.
Discussion
This study is among the first to examine changes in global gene expression in chondrocytes after nuclear receptor agonist treatment, particularly paired with concurrent functional analysis. It provides compelling evidence that nuclear receptors drive early changes in cell metabolism that can influence deleterious changes in cellular phenotype leading to the progression of OA. Nuclear receptors have been increasingly linked to the progression of OA. We have previously established the degenerative changes promoted by PPARδ agonism in cartilage, as well as the beneficial and necessary role of PPARγ in cartilage [13, 14]. We and others have characterized the protective role of LXR in osteoarthritis [16, 35, 36]. However, in order to establish how or whether these ligand-activated receptors are feasible therapeutic targets, we must examine the molecular changes linked to activation or inhibition of each factor.
We used IMACs treated with LXR, RXR, PPARγ, or PPARδ agonists for 72 h. Immature murine articular chondrocytes provide a large number of cells for analyses on fully differentiated primary chondrocytes while minimizing dedifferentiation [19]. Microarray analyses of IMACs revealed changes in metabolic and ECM genes in response to these agonists; these changes were largely confirmed by qPCR. Agonism of RXR decreased gene expression of the major ECM component aggrecan and increased the expression of ECM protease Mmp13, while LXR agonism decreased the gene expression of proteases Adamts4, Mmp2, and Mmp13. Of particular interest were the increases in expression of genes involved in lipid metabolism since they showed greater induction than those regulating ECM turnover. Among these genes, two were induced by all four agonists, Pdk4 and Angptl4, suggesting that they might play central roles in cartilage metabolism. Interestingly, in an earlier study, we had also demonstrated increased expression of Pdk4 in response to dexamethasone, a ligand for the glucocorticoid receptor which is another member of the nuclear receptor family [37].
Functional evaluation of lipid metabolism using cellular lipid mass assays demonstrated a significant decrease in triglycerides after PPARδ agonist treatment. Conversely, triglycerides were significantly increased with PPARγ, LXR, and RXR agonists. This is not surprising as PPARγ can often act antagonistically to PPARδ with regard to lipogenesis [38], while LXR mediates fatty acid biosynthesis through activation of genes such as Srebf1, Fasn, and Scd1 which corroborates our data (see GEO dataset) [39, 40]. Quantification of cell lipids in vitro enables us to assess differences in some aspects of lipid metabolism between treatments. In fact, it is plausible that the dysregulation in lipid metabolism that we observed could initiate metabolic changes in the cell that eventually lead to apoptosis, inflammation, or changes in cell behavior, such as synthesis of catabolic factors. Increased lipid deposition in osteoarthritic cartilage has been shown, while increased reactive oxygen species (ROS) can cause lipid peroxidation, which in turn could cause oxidative stress, resulting in degenerative changes to the matrix through oxidation of collagen II [6]. We also sought to reduce the dysregulation of cell lipids through the pharmacological inhibition of PDK, which would increase ATP synthesis through glucose oxidation [41]. However, we found that further attempting to shift the preferred metabolic pathway away from fatty acid oxidation by promoting an alternate pathway did not change cellular triglyceride levels and agonist responses.
In addition to dysregulation of cell lipids, we also see significantly decreased Gsta4 expression after PPARδ or RXR agonist treatment. The encoded enzyme Glutathione S-transferase A 4 protects against HNE (4 Hydroxynonal)-induced damage in chondrocytes. HNE is an extremely reactive aldehyde produced from ROS and lipid peroxidation and is increased in synovial fluid from OA patients [40] as well as articular cartilage [42]. HNE can also post-transcriptionally modify Collagen 2 and MMP13 to induce degradative changes in cartilage as well as changes in cell–matrix interactions [43, 44]. Molecularly, it has been observed that HNE can stimulate COX-2 via the ATF/cAMP response element and inhibits iNOS and NF-kB inactivation in human articular chondrocytes [45]. GSTs are major determinants of intracellular HNE concentration, and dysregulation in disease states can result in toxic effects [46]. The GSTA4 isoform has well-established selectivity and efficiency for conjugation of lipid peroxidation products, especially HNE [46, 47]. Accordingly, decreased levels of GSTA4 are present in human OA cartilage, making the cartilage more susceptible to damage [33]. In addition, IL-1β treatment of human articular chondrocytes decreases expression of GSTA4, while inhibition of the p38 MAPK pathway (an important signaling pathway in the pathogenesis of OA) increases levels of GSTA4 [48]. This suggests that the catabolic effects of PPARδ (and possibly RXR) in cartilage could be partially due to the decreased expression of Gsta4, resulting in decreased anti-oxidant defense and thus increased oxidative stress. In our surgical model of DMM induction, we see that even the minimal damage experienced by PPARδ KO after surgery is enough to decrease the expression of this protein in vivo, indicating that Gsta4 is an early response gene to damage and that there may be multiple pathways that influence its regulation.
Another gene of interest, Txnip (thioredoxin interacting protein), was highly induced by PPARδ agonism, but appeared repressed by LXR agonism, in agreement with the opposing effects of these nuclear receptors on OA progression. Thioredoxin is another important antioxidant, but binding of Txnip to thioredoxin inhibits its ability to scavenge for ROS [49]. In humans, decreased TXNIP is seen at late stages of OA, while increased TXNIP is seen in H2O2-induced cartilage damage in human chondrocytes [50, 51]. In our study, we demonstrate increased gene expression of Txnip after PPARδ agonism in chondrocytes, and control mice show increased staining for Txnip after DMM surgery. The apparent differences in Txnip regulation in human and mouse OA are likely due to the different disease stages; human data were obtained from end-stage samples, while the DMM model is subtle and our analyses have largely focused on mild to moderate disease. We also show that cartilage-specific Ppard knockout mice that are protected against cartilage degeneration in the DMM model have decreased Txnip staining after DMM surgery, in line with our microarray data. Txnip has been linked to oxidative stress and inflammation, and it can directly activate NF-kB and downstream inflammatory cytokines [52]. In chondrocytes, recent work has shown that Redd1 can form a complex with Txnip to regulate autophagy [53]. Taken altogether, these data help to form a cohesive picture of how changes in cell metabolism could influence the development of early osteoarthritis. Nuclear receptors appear to play a key role in these processes by regulating the expression of central players such as Txnip and Gsta4.
Current treatment strategies for OA are largely ineffective or inconclusive. It is possible that we are missing a critical temporal period during which chondrocyte homeostasis is disrupted, later leading to matrix degeneration. Recent evidence demonstrates that nuclear receptors are key regulators of OA pathogenesis, and the data presented here suggests that their primary targets are metabolic regulation. Metabolic deregulation, in turn, can trigger events leading to oxidative stress and inflammation, protease activation, and ultimately cartilage degeneration. Targeting these critical processes could be a promising avenue for treatments that alter disease progression.
References
Neogi T, Zhang Y (2013) Epidemiology of osteoarthritis. Rheum Dis Clin N Am 39:1–19
Moskowitz RW (1999) Bone remodeling in osteoarthritis: subchondral and osteophytic responses. Osteoarthr Cartil 7:323–324
Fitzgerald JB, Jin M, Dean D, Wood DJ, Zheng MH, Grodzinsky AJ (2004) Mechanical compression of cartilage explants induces multiple time-dependent gene expression patterns and involves intracellular calcium and cyclic AMP. J Biol Chem 279:19502–19511
Zhuo Q, Yang W, Chen J, Wang Y (2012) Metabolic syndrome meets osteoarthritis. Nat Rev Rheumatol 8:729–737
Masuko K, Murata M, Suematsu N, Okamoto K, Yudoh K, Nakamura H, Kato T (2009) A metabolic aspect of osteoarthritis: lipid as a possible contributor to the pathogenesis of cartilage degradation. Clin Exp Rheumatol 27:347–353
Lippiello L, Walsh T, Fienhold M (1991) The association of lipid abnormalities with tissue pathology in human osteoarthritic articular cartilage. Metabolism 40:571–576
Ali SA, Al-Jazrawe M, Ma H, Whetstone H, Poon R, Farr S, Naples M, Adeli K, Alman BA (2016) Regulation of cholesterol homeostasis by hedgehog signaling in osteoarthritic cartilage. Arthritis Rheumatol 68:127–137
Griffin TM, Huebner JL, Kraus VB, Yan Z, Guilak F (2012) Induction of osteoarthritis and metabolic inflammation by a very high-fat diet in mice: effects of short-term exercise. Arthritis Rheum 64:443–453
Wu CL, Jain D, McNeill JN, Little D, Anderson JA, Huebner JL, Kraus VB, Rodriguiz RM, Wetsel WC, Guilak F (2015) Dietary fatty acid content regulates wound repair and the pathogenesis of osteoarthritis following joint injury. Ann Rheum Dis 74:2076–2083
Bourguet W, Germain P, Gronemeyer H (2000) Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci 21:381–388
Rangwala SM, Lazar MA (2004) Peroxisome proliferator-activated receptor gamma in diabetes and metabolism. Trends Pharmacol Sci 25:331–336
Beaven SW, Tontonoz P (2006) Nuclear receptors in lipid metabolism: targeting the heart of dyslipidemia. Annu Rev Med 57:313–329
Ratneswaran A, LeBlanc EA, Walser E, Welch I, Mort JS, Borradaile N, Beier F (2015) Peroxisome proliferator-activated receptor delta promotes the progression of posttraumatic osteoarthritis in a mouse model. Arthritis Rheumatol 67:454–464
Vasheghani F, Monemdjou R, Fahmi H, Zhang Y, Perez G, Blati M, St-Arnaud R, Pelletier JP, Beier F, Martel-Pelletier J et al (2013) Adult cartilage-specific peroxisome proliferator-activated receptor gamma knockout mice exhibit the spontaneous osteoarthritis phenotype. Am J Pathol 182:1099–1106
Monemdjou R, Vasheghani F, Fahmi H, Perez G, Blati M, Taniguchi N, Lotz M, St-Arnaud R, Pelletier JP, Martel-Pelletier J et al (2012) Association of cartilage-specific deletion of peroxisome proliferator-activated receptor gamma with abnormal endochondral ossification and impaired cartilage growth and development in a murine model. Arthritis Rheum 64:1551–1561
Sun MM, Beier F (2014) Liver X receptor activation delays chondrocyte hypertrophy during endochondral bone growth. Osteoarthr Cartil 22:996–1006
Bando Y, Yamamoto M, Sakiyama K, Inoue K, Takizawa S, Owada Y, Iseki S, Kondo H, Amano O (2014) Expression of epidermal fatty acid binding protein (E-FABP) in septoclasts in the growth plate cartilage of mice. J Mol Histol 45:507–518
Collins-Racie LA, Yang Z, Arai M, Li N, Majumdar MK, Nagpal S, Mounts WM, Dorner AJ, Morris E, LaVallie ER (2009) Global analysis of nuclear receptor expression and dysregulation in human osteoarthritic articular cartilage: reduced LXR signaling contributes to catabolic metabolism typical of osteoarthritis. Osteoarthr Cartil 17:832–842
Salvat C, Pigenet A, Humbert L, Berenbaum F, Thirion S (2005) Immature murine articular chondrocytes in primary culture: a new tool for investigating cartilage. Osteoarthr Cartil 13:243–249
McCann MR, Patel P, Pest MA, Ratneswaran A, Lalli G, Beaucage KL, Backler GB, Kamphuis MP, Esmail Z, Lee J et al (2015) Repeated exposure to high-frequency low-amplitude vibration induces degeneration of murine intervertebral discs and knee joints. Arthritis Rheumatol 67:2164–2175
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408
Appleton CT, Usmani SE, Mort JS, Beier F (2010) Rho/ROCK and MEK/ERK activation by transforming growth factor-alpha induces articular cartilage degradation. Lab Investig 90:20–30
Appleton CT, Pitelka V, Henry J, Beier F (2007) Global analyses of gene expression in early experimental osteoarthritis. Arthritis Rheum 56:1854–1868
Oliveros JC (2007–2015) Venny. An interactive tool for comparing lists with Venn’s diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html
Whitman SC, Sawyez CG, Miller DB, Wolfe BM, Huff MW (1998) Oxidized type IV hypertriglyceridemic VLDL-remnants cause greater macrophage cholesteryl ester accumulation than oxidized LDL. J Lipid Res 39:1008–1020
Pest MA, Russell BA, Zhang YW, Jeong JW, Beier F (2014) Disturbed cartilage and joint homeostasis resulting from a loss of mitogen-inducible gene 6 in a mouse model of joint dysfunction. Arthritis Rheumatol 66:2816–2827
Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J (2008) Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab 8:347–358
Mandard S, Zandbergen F, Tan NS, Escher P, Patsouris D, Koenig W, Kleemann R, Bakker A, Veenman F, Wahli W et al (2004) The direct peroxisome proliferator-activated receptor target fasting-induced adipose factor (FIAF/PGAR/ANGPTL4) is present in blood plasma as a truncated protein that is increased by fenofibrate treatment. J Biol Chem 279:34411–34420
Hermann LM, Pinkerton M, Jennings K, Yang L, Grom A, Sowders D, Kersten S, Witte DP, Hirsch R, Thornton S (2005) Angiopoietin-like-4 is a potential angiogenic mediator in arthritis. Clin Immunol 115:93–101
Mathieu M, Iampietro M, Chuchana P, Guerit D, Djouad F, Noel D, Jorgensen C (2014) Involvement of angiopoietin-like 4 in matrix remodeling during chondrogenic differentiation of mesenchymal stem cells. J Biol Chem 289:8402–8412
Murata M, Yudo K, Nakamura H, Chiba J, Okamoto K, Suematsu N, Nishioka K, Beppu M, Inoue K, Kato T et al (2009) Hypoxia upregulates the expression of angiopoietin-like-4 in human articular chondrocytes: role of angiopoietin-like-4 in the expression of matrix metalloproteinases and cartilage degradation. J Orthop Res 27:50–57
Lee J, Ozcan U (2014) Unfolded protein response signaling and metabolic diseases. J Biol Chem 289:1203–1211
Vaillancourt F, Fahmi H, Shi Q, Lavigne P, Ranger P, Fernandes JC, Benderdour M (2008) 4-hydroxynonenal induces apoptosis in human osteoarthritic chondrocytes: the protective role of glutathione-S-transferase. Arthritis Res Ther 10:R107
Nakamura MT, Yudell BE, Loor JJ (2014) Regulation of energy metabolism by long-chain fatty acids. Prog Lipid Res 53:124–144
Li N, Rivera-Bermudez MA, Zhang M, Tejada J, Glasson SS, Collins-Racie LA, Lavallie ER, Wang Y, Chang KC, Nagpal S et al (2010) LXR modulation blocks prostaglandin E2 production and matrix degradation in cartilage and alleviates pain in a rat osteoarthritis model. Proc Natl Acad Sci U S A 107:3734–3739
Tsezou A, Iliopoulos D, Malizos KN, Simopoulou T (2010) Impaired expression of genes regulating cholesterol efflux in human osteoarthritic chondrocytes. J Orthop Res 28:1033–1039
James CG, Ulici V, Tuckermann J, Underhill TM, Beier F (2007) Expression profiling of dexamethasone-treated primary chondrocytes identifies targets of glucocorticoid signalling in endochondral bone development. BMC Genomics 8:205
Roberts LD, Murray AJ, Menassa D, Ashmore T, Nicholls AW, Griffin JL (2011) The contrasting roles of PPARdelta and PPARgamma in regulating the metabolic switch between oxidation and storage of fats in white adipose tissue. Genome Biol 12:R75
Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ et al (2000) Role of LXRs in control of lipogenesis. Genes Dev 14:2831–2838
Morquette B, Shi Q, Lavigne P, Ranger P, Fernandes JC, Benderdour M (2006) Production of lipid peroxidation products in osteoarthritic tissues: new evidence linking 4-hydroxynonenal to cartilage degradation. Arthritis Rheum 54:271–281
Piao L, Fang Y-H, Cadete VJJ, Wietholt C, Urboniene D, Toth PT, Marsboom G, Zhang HJ, Haber I, Rehman J et al (2009) The inhibition of pyruvate dehydrogenase kinase improves impaired cardiac function and electrical remodeling in two models of right ventricular hypertrophy: resuscitating the hibernating right ventricle. J Mol Med 88:47–60
Shah R, Raska K Jr, Tiku ML (2005) The presence of molecular markers of in vivo lipid peroxidation in osteoarthritic cartilage: a pathogenic role in osteoarthritis. Arthritis Rheum 52:2799–2807
El-Bikai R, Welman M, Margaron Y, Cote JF, Macqueen L, Buschmann MD, Fahmi H, Shi Q, Maghni K, Fernandes JC et al (2010) Perturbation of adhesion molecule-mediated chondrocyte-matrix interactions by 4-hydroxynonenal binding: implication in osteoarthritis pathogenesis. Arthritis Res Ther 12:R201
Bentz M, Zaouter C, Shi Q, Fahmi H, Moldovan F, Fernandes JC, Benderdour M (2012) Inhibition of inducible nitric oxide synthase prevents lipid peroxidation in osteoarthritic chondrocytes. J Cell Biochem 113:2256–2267
Vaillancourt F, Morquette B, Shi Q, Fahmi H, Lavigne P, Di Battista JA, Fernandes JC, Benderdour M (2007) Differential regulation of cyclooxygenase-2 and inducible nitric oxide synthase by 4-hydroxynonenal in human osteoarthritic chondrocytes through ATF-2/CREB-1 transactivation and concomitant inhibition of NF-kappaB signaling cascade. J Cell Biochem 100:1217–1231
Balogh LM, Atkins WM (2011) Interactions of glutathione transferases with 4-hydroxynonenal. Drug Metab Rev 43:165–178
Ruskovska T, Bernlohr DA (2013) Oxidative stress and protein carbonylation in adipose tissue - implications for insulin resistance and diabetes mellitus. J Proteome 92:323–334
Joos H, Albrecht W, Laufer S, Brenner RE (2009) Influence of p38MAPK inhibition on IL-1beta-stimulated human chondrocytes: a microarray approach. Int J Mol Med 23:685–693
Nishiyama A, Matsui M, Iwata S, Hirota K, Masutani H, Nakamura H, Takagi Y, Sono H, Gon Y, Yodoi J (1999) Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J Biol Chem 274:21645–21650
Karlsson C, Dehne T, Lindahl A, Brittberg M, Pruss A, Sittinger M, Ringe J (2010) Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthr Cartil 18:581–592
Lim HD, Kim YS, Ko SH, Yoon IJ, Cho SG, Chun YH, Choi BJ, Kim EC (2012) Cytoprotective and anti-inflammatory effects of melatonin in hydrogen peroxide-stimulated CHON-001 human chondrocyte cell line and rabbit model of osteoarthritis via the SIRT1 pathway. J Pineal Res 53:225–237
Mohamed IN, Hafez SS, Fairaq A, Ergul A, Imig JD, El-Remessy AB (2014) Thioredoxin-interacting protein is required for endothelial NLRP3 inflammasome activation and cell death in a rat model of high-fat diet. Diabetologia 57:413–423
Alvarez-Garcia O, Olmer M, Akagi R, Akasaki Y, Fisch KM, Shen T, Su AI, Lotz MK (2016) Suppression of REDD1 in osteoarthritis cartilage, a novel mechanism for dysregulated mTOR signaling and defective autophagy. Osteoarthr Cartil. doi:10.1016/j.joca.2016.04.015
Acknowledgements
A.R. and M.M.-G.S. were supported by Doctoral Scholarships from The Arthritis Society (Canada) and the Collaborative Training Program in Musculoskeletal Health Research at The University of Western Ontario. M.M.-G.S. is also supported by the Dean’s Transfer Stipend. F.B. holds a Canada Research Chair in Musculoskeletal Health Research. This work was supported by operating grants from the Canadian Institutes of Health Research and The Arthritis Society (Canada). We thank all members of the Beier lab for discussions and support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests related to this study.
Additional information
Anusha Ratneswaran and Margaret Man-Ger Sun contributed equally.
Electronic supplementary material
.
ESM 1
(PDF 3578 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ratneswaran, A., Sun, M.MG., Dupuis, H. et al. Nuclear receptors regulate lipid metabolism and oxidative stress markers in chondrocytes. J Mol Med 95, 431–444 (2017). https://doi.org/10.1007/s00109-016-1501-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-016-1501-5