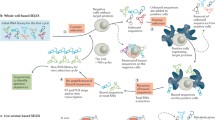
Abstract
Aptamers are single-stranded oligonucleotides that fold into well-defined three-dimensional shapes, allowing them to bind their targets with high affinity and specificity. They can be generated through an in vitro process called “Systemic Evolution of Ligands by Exponential Enrichment” and applied for specific detection, inhibition, and characterization of various targets like small organic and inorganic molecules, proteins, and whole cells. Aptamers have also been called chemical antibodies because of their synthetic origin and their similar modes of action to antibodies. They exhibit significant advantages over antibodies in terms of their small size, synthetic accessibility, and ability to be chemically modified and thus endowed with new properties. The first generation of aptamer drug “Macugen” was available for public use within 25 years of the discovery of aptamers. With others in the pipeline for clinical trials, this emerging field of medical biotechnology is raising significant interest. However, aptamers pose different problems for their development than for antibodies that need to be addressed to achieve practical applications. It is likely that current developments in aptamer engineering will be the basis for the evolution of improved future bioanalytical and biomedical applications. The present review discusses the development of aptamers for therapeutics, drug delivery, target validation and imaging, and reviews some of the challenges to fully realizing the promise of aptamers in biomedical applications.


Similar content being viewed by others
References
Hermann T, Patel DJ (2000) Adaptive recognition by nucleic acid aptamers. Science 287:820–825
Zhang Q, Sun X, Watt ED, Al-Hashimi HM (2006) Resolving the motional modes that code for RNA adaptation. Science 311:653–656
Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM (2007) Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature 450:1263–1267
Duchardt-Ferner E, Weigand JE, Ohlenschlager O, Schmidtke SR, Suess B, Wohnert J (2010) Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch. Angew Chem Int Ed Engl 49:6216–6219
Wunnicke D, Strohbach D, Weigand JE, Appel B, Feresin E, Suess B, Muller S, Steinhoff HJ (2011) Ligand-induced conformational capture of a synthetic tetracycline riboswitch revealed by pulse EPR. RNA 17:182–188
Chen B, Zuo X, Wang YX, Dayie TK (2012) Multiple conformations of SAM-II riboswitch detected with SAXS and NMR spectroscopy. Nucleic Acids Res 40:3117–3130
Banerjee J (2010) Antibodies are challenged. Indian J Med Sci 64:144–147
Jayasena S (1999) Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem 45:1628–1650
Lo AS, Zhu Q, Marasco WA (2008) Intracellular antibodies (intrabodies) and their therapeutic potential. Handb Exp Pharmacol 181:343–373
Que-Gewirth NS, Sullenger BA (2007) Gene therapy progress and prospects: RNA aptamers. Gene Ther 14:283–291
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822
Robertson DL, Joyce GF (1990) Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344:467–468
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Mi J, Liu Y, Rabbani ZN, Yang Z, Urban JH, Sullenger BA, Clary BM (2010) In vivo selection of tumor-targeting RNA motifs. Nat Chem Biol 6:22–24
Cheng C, Chen Y, Lennox K, Behlke M, Davidson BL (2013) In vivo SELEX for identification of brain-penetrating aptamers. Mol Ther Nucleic Acids 2:e67. doi:10.1038/mt.2012.59
Cox JC, Ellington AD (2001) Automated selection of anti-protein aptamers. Bioorg Med Chem 9:2525–2531
Lee JF, Stovall GM, Ellington AD (2006) Aptamer therapeutics advance. Curr Opin Chem Biol 10:282–289
Kusser W (2000) Chemically modified nucleic acid aptamers for in vitro selections: evolving evolution. J Biotechnol 74:27–38
Foy JW, Rittenhouse K, Modi M, Patel M (2007) Local tolerance and systemic safety of pegaptanib sodium in the dog and rabbit. J Ocul Pharmacol Ther 23:452–466
Yu D, Wang D, Zhu FG, Bhagat L, Dai M, Kandimalla ER, Agrawal S (2009) Modifications incorporated in CpG motifs of oligodeoxynucleotides lead to antagonist activity of toll-like receptors 7 and 9. J Med Chem 52:5108–5114
Ng EW, Adamis AP (2006) Anti-VEGF aptamer (pegaptanib) therapy for ocular vascular diseases. Ann N Y Acad Sci 1082:151–171
Gold L, Janjic N, Jarvis T, Schneider D, Walker JJ, Wilcox SK, Zichi D (2012) Aptamers and the RNA world, past and present. Cold Spring Harb Perspect Biol 4:a003582
Sundaram P, Kurniawan H, Byrne M, Wower J (2013) Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci 48:259–271
DeAnda A Jr, Coutre SE, Moon MR, Vial CM, Griffin LC, Law VS, Komeda M, Leung LL, Miller DC (1994) Pilot study of the efficacy of a thrombin inhibitor for use during cardiopulmonary bypass. Ann Thorac Surg 58:344–350
Rusconi CP, Roberts JD, Pitoc GA, Nimjee SM, White RR, Quick G Jr, Scardino E, Fay WP, Sullenger BA (2004) Antidote-mediated control of an anticoagulant aptamer in vivo. Nat Biotechnol 22:1423–1428
Blind M, Kolanus W, Famulok M (1999) Cytoplasmic RNA modulators of an inside-out signal-transduction cascade. PNAS 96:3606–3610
DeStefano JJ, Nair GR (2008) Novel aptamer inhibitors of human immunodeficiency virus reverse transcriptase. Oligonucleotides 18:133–144
Li N, Wang Y, Pothukuchy A, Syrett A, Husain N, Gopalakrisha S, Kosaraju P, Ellington AD (2008) Aptamers that recognize drug-resistant HIV-1 reverse transcriptase. Nucleic Acids Res 36:6739–6751
Michalowski D, Chitima-Matsiga R, Held DM, Burke DH (2008) Novel bimodular DNA aptamers with guanosine quadruplexes inhibit phylogenetically diverse HIV-1 reverse transcriptases. Nucleic Acids Res 36:7124–7135
Ditzler MA, Bose D, Shkriabai N, Marchand B, Sarafianos SG, Kvaratskhelia M, Burke DH (2011) Broad-spectrum aptamer inhibitors of HIV reverse transcriptase closely mimic natural substrates. Nucleic Acids Res 39:8237–8247
Symensma TL, Giver L, Zapp M, Takle GB, Ellington AD (1996) RNA aptamers selected to bind human immunodeficiency virus type 1 Rev in vitro are Rev responsive in vivo. J Virol 70:179–187
Yamamoto R, Toyoda S, Viljanen P, Machida K, Nishikawa S, Murakami K, Taira K, Kumar PK (1995) In vitro selection of RNA aptamers that can bind specifically to Tat protein of HIV-1. Nucleic Acids Symp Ser 34:145–146
Allen P, Worland S, Gold L (1995) Isolation of high-affinity RNA ligands to HIV-1 integrase from a random pool. Virology 209:327–336
Kohn DB, Bauer G, Rice CR, Rothschild JC, Carbonaro DA, Valdez P, Q-l H, Zhou C, Bahner I, Kearns K et al (1999) A Clinical trial of retroviral-mediated transfer of arev-responsive element decoy gene into CD34 + Cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood 94:368–371
Dey AK, Khati M, Tang M, Wyatt R, Lea SM, James W (2005) An aptamer that neutralizes R5 strains of human immunodeficiency virus type 1 blocks gp120-CCR5 interaction. J Virol 79:13806–13810
Cohen C, Forzan M, Sproat B, Pantophlet R, McGowan I, Burton D, James W (2008) An aptamer that neutralizes R5 strains of HIV-1 binds to core residues of gp120 in the CCR5 binding site. Virology 381:46–54
Mufhandu HT, Gray ES, Madiga MC, Tumba N, Alexandre KB, Khoza T, Wibmer CK, Moore PL, Morris L, Khati M (2012) UCLA1, a synthetic derivative of a gp120 RNA aptamer, inhibits entry of human immunodeficiency virus type 1 subtype C. J Virol 86:4989–4999
Neff CP, Zhou J, Remling L, Kuruvilla J, Zhang J, Li H, Smith DD, Swiderski P, Rossi JJ, Akkina R (2011) An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci Transl Med 3:66ra66. doi:10.1126/scitranslmed.3001581
Binning J, Leung D, Amarasinghe G (2012) Aptamers in virology: recent advances and challenges. Front Microbiol 3:29
Adler A, Forster N, Homann M, Goringer HU (2008) Post-SELEX chemical optimization of a trypanosome-specific RNA aptamer. Comb Chem High Throughput Screen 11:16–23
Barfod A, Persson T, Lindh J (2009) In vitro selection of RNA aptamers against a conserved region of the Plasmodium falciparum erythrocyte membrane protein 1. Parasitol Res 105:1557–1566
Chen F, Zhang X, Zhou J, Liu S, Liu J (2012) Aptamer inhibits Mycobacterium tuberculosis (H37Rv) invasion of macrophage. Mol Biol Rep 39:2157–2162
Li H, Ding X, Peng Z, Deng L, Wang D, Chen H, He Q (2011) Aptamer selection for the detection of Escherichia coli K88. Can J Microbiol 57:453–459
Torres-Chavolla E, Alocilja EC (2009) Aptasensors for detection of microbial and viral pathogens. Biosens Bioelectron 24:3175–3182
Sefah K, Tang Z, Shangguan D, Chen H, Lopez-Colon D, Li Y, Parekh P, Martin J, Meng L, Phillips J et al (2009) Molecular recognition of acute myeloid leukemia using aptamers. Leukemia 23:235–244
Ye M, Hu J, Peng M, Liu J, Liu J, Liu H, Zhao X, Tan W (2012) Generating aptamers by cell-SELEX for applications in molecular medicine. Int J Mol Sci 13:3341–3353
Graham J, Zarbl H (2012) Use of cell-SELEX to generate DNA aptamers as molecular probes of HPV-associated cervical cancer cells. PLoS ONE 7:e36103
Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO (2009) Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol 86:151–164
Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ (2008) The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res 68:2358–2365
Reyes-Reyes EM, Teng Y, Bates PJ (2010) A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res 70:8617–8629
Mi Z, Guo H, Russell MB, Liu Y, Sullenger BA, Kuo PC (2009) RNA aptamer blockade of osteopontin inhibits growth and metastasis of MDA-MB231 breast cancer cells. Mol Ther 17:153–161
Blake CM, Sullenger BA, Lawrence DA, Fortenberry YM (2009) Antimetastatic potential of PAI-1-specific RNA aptamers. Oligonucleotides 19:117–128
Liu Y, Kuan CT, Mi J, Zhang X, Clary BM, Bigner DD, Sullenger BA (2009) Aptamers selected against the unglycosylated EGFRvIII ectodomain and delivered intracellularly reduce membrane-bound EGFRvIII and induce apoptosis. Biol Chem 390:137–144
Ferreira CS, Cheung MC, Missailidis S, Bisland S, Gariepy J (2009) Phototoxic aptamers selectively enter and kill epithelial cancer cells. Nucleic Acids Res 37:866–876
Heldin CH, Rubin K, Pietras K, Ostman A (2004) High interstitial fluid pressure—an obstacle in cancer therapy. Nat Rev Cancer 4:806–813
Pendergrast PS, Marsh HN, Grate D, Healy JM, Stanton M (2005) Nucleic acid aptamers for target validation and therapeutic applications. J Biomol Tech 16:224–234
Takemura K, Wang P, Vorberg I, Surewicz W, Priola SA, Kanthasamy A, Pottathil R, Chen SG, Sreevatsan S (2006) DNA aptamers that bind to PrP(C) and not PrP(Sc) show sequence and structure specificity. Exp Biol Med (Maywood) 231:204–214
Gilch S, Schatzl HM (2009) Aptamers against prion proteins and prions. Cell Mol Life Sci 66:2445–2455
Hwang B, Han K, Lee SW (2003) Prevention of passively transferred experimental autoimmune myasthenia gravis by an in vitro selected RNA aptamer. FEBS Lett 548:85–89
Gutsaeva DR, Parkerson JB, Yerigenahally SD, Kurz JC, Schaub RG, Ikuta T, Head CA (2011) Inhibition of cell adhesion by anti-P-selectin aptamer: a new potential therapeutic agent for sickle cell disease. Blood 117:727–735
Haile LA, von Wasielewski R, Gamrekelashvili J, Krüger C, Bachmann O, Westendorf AM, Buer J, Liblau R, Manns MP, Korangy F et al (2008) Myeloid-derived suppressor cells in inflammatory bowel disease: a new immunoregulatory pathway. Gastroenterology 135:871–881. doi:10.1053/j.gastro.2008.06.032, e5
Waldron T, Quatromoni J, Karakasheva T, Singhal S, Rustgi A (2013) Myeloid-derived suppressor cells: targets for therapy. Oncol Immunol 2:e24117
Roth F, De La Fuente AC, Vella JL, Zoso A, Inverardi L, Serafini P (2012) Aptamer-mediated blockade of IL4Rα triggers apoptosis of MDSCs and limits tumor progression. Cancer Res 72(6):1373–1383
Ray P, White R (2010) Aptamers for targeted drug delivery. Pharmaceuticals 3:1761–1778
Meyer C, Hahn U, Rentmeister A (2011) Cell-specific aptamers as emerging therapeutics. J Nucleic Acids 2011:904750
Bagalkot V, Zhang L, Levy-Nissenbaum E, Jon S, Kantoff PW, Langer R, Farokhzad OC (2007) Quantum dot-aptamer conjugates for synchronous cancer imaging, therapy, and sensing of drug delivery based on bi-fluorescence resonance energy transfer. Nano Lett 7:3065–3070
Zhang L, Radovic-Moreno AF, Alexis F, Gu FX, Basto PA, Bagalkot V, Jon S, Langer RS, Farokhzad OC (2007) Co-delivery of hydrophobic and hydrophilic drugs from nanoparticle-aptamer bioconjugates. Chem Med Chem 2:1268–1271
Xiao Z, Shangguan D, Cao Z, Fang X, Tan W (2008) Cell-specific internalization study of an aptamer from whole cell selection. Chemistry 14:1769–1775
McNamara JO 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH (2006) Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol 24:1005–1015
Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO 2nd, Giangrande PH (2009) Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol 27:839–849
Chu TC, Twu KY, Ellington AD, Levy M (2006) Aptamer mediated siRNA delivery. Nucleic Acids Res 34:e73
Wullner U, Neef I, Eller A, Kleines M, Tur MK, Barth S (2008) Cell-specific induction of apoptosis by rationally designed bivalent aptamer-siRNA transcripts silencing eukaryotic elongation factor 2. Curr Cancer Drug Targets 8:554–565
Chu TC, Marks JW 3rd, Lavery LA, Faulkner S, Rosenblum MG, Ellington AD, Levy M (2006) Aptamer: toxin conjugates that specifically target prostate tumor cells. Cancer Res 66:5989–5992
Zhang K, Sefah K, Tang L, Zhao Z, Zhu G, Ye M, Sun W, Goodison S, Tan W (2012) A novel aptamer developed for breast cancer cell internalization. Chem Med Chem 7:79–84
Chen CH, Dellamaggiore KR, Ouellette CP, Sedano CD, Lizadjohry M, Chernis GA, Gonzales M, Baltasar FE, Fan AL, Myerowitz R et al (2008) Aptamer-based endocytosis of a lysosomal enzyme. Proc Natl Acad Sci U S A 105:15908–15913
Tong GJ, Hsiao SC, Carrico ZM, Francis MB (2009) Viral capsid DNA aptamer conjugates as multivalent cell-targeting vehicles. J Am Chem Soc 131:11174–11178
Huang YF, Chang HT, Tan W (2008) Cancer cell targeting using multiple aptamers conjugated on nanorods. Anal Chem 80:567–572
Wu Y, Phillips JA, Liu H, Yang R, Tan W (2008) Carbon nanotubes protect DNA strands during cellular delivery. ACS Nano 2:2023–2028
Kang H, O'Donoghue MB, Liu H, Tan W (2010) A liposome-based nanostructure for aptamer directed delivery. Chem Commun (Camb) 46:249–251
Cao Z, Tong R, Mishra A, Xu W, Wong GC, Cheng J, Lu Y (2009) Reversible cell-specific drug delivery with aptamer-functionalized liposomes. Angew Chem Int Ed Engl 48:6494–6498
Choi JH, Chen KH, Strano MS (2006) Aptamer-capped nanocrystal quantum dots: a new method for label-free protein detection. J Am Chem Soc 128:15584–15585
Park JU, Lee JH, Paik U, Lu Y, Rogers JA (2008) Nanoscale patterns of oligonucleotides formed by electrohydrodynamic jet printing with applications in biosensing and nanomaterials assembly. Nano Lett 8:4210–4216
Levy M, Cater SF, Ellington AD (2005) Quantum-dot aptamer beacons for the detection of proteins. Chem Biochem 6:2163–2166
Wernette DP, Liu J, Bohn PW, Lu Y (2008) Functional-DNA-based nanoscale materials and devices for sensing trace contaminants in water. MRS Bull 33:34–41
Li H, Wang C, Wu Z, Lu L, Qiu L, Zhou H, Shen G, Yu R (2013) An electronic channel switching-based aptasensor for ultrasensitive protein detection. Anal Chim Acta 758:130–137
Zhai L, Wang T, Kang K, Zhao Y, Shrotriya P, Nilsen-Hamilton M (2012) An RNA aptamer-based microcantilever sensor to detect the inflammatory marker, mouse lipocalin-2. Anal Chem 84:8763–8770
Swearingen CB, Wernette DP, Cropek DM, Lu Y, Sweedler JV, Bohn PW (2005) Immobilization of a catalytic DNA molecular beacon on Au for Pb(II) detection. Anal Chem 77:442–448
Du Y, Chen C, Zhou M, Dong S, Wang E (2011) Microfluidic electrochemical aptameric assay integrated on-chip: a potentially convenient sensing platform for the amplified and multiplex analysis of small molecules. Anal Chem 83:1523–1529
Mehan MR, Ostroff R, Wilcox SK, Steele F, Schneider D, Jarvis TC, Baird GS, Gold L, Janjic N (2013) Highly multiplexed proteomic platform for biomarker discovery, diagnostics, and therapeutics. Adv Exp Med Biol 734:283–300
Brody E, Gold L, Mehan M, Ostroff R, Rohloff J, Walker J, Zichi D (2012) Life's simple measures: unlocking the proteome. J Mol Biol 422:595–606
Liu J, Lee JH, Lu Y (2007) Quantum dot encoding of aptamer-linked nanostructures for one-pot simultaneous detection of multiple analytes. Anal Chem 79:4120–4125
Kuo TC, Cannon DM Jr, Chen Y, Tulock JJ, Shannon MA, Sweedler JV, Bohn PW (2003) Gateable nanofluidic interconnects for multilayered microfluidic separation systems. Anal Chem 75:1861–1867
Xu Y, Phillips JA, Yan J, Li Q, Fan ZH, Tan W (2009) Aptamer-based microfluidic device for enrichment, sorting, and detection of multiple cancer cells. Anal Chem 81:7436–7442
Stojanovic MN, Landry DW (2002) Aptamer-based colorimetric probe for cocaine. J Am Chem Soc 124:9678–9679
Wang Y, Li D, Ren W, Liu Z, Dong S, Wang E (2008) Ultrasensitive colorimetric detection of protein by aptamer-Au nanoparticles conjugates based on a dot-blot assay. Chem Commun (Camb) 44:2520–2522
Wang L, Liu X, Hu X, Song S, Fan C (2006) Unmodified gold nanoparticles as a colorimetric probe for potassium DNA aptamers. Chem Commun (Camb) 36:3780–3782
Wei H, Li B, Li J, Wang E, Dong S (2007) Simple and sensitive aptamer-based colorimetric sensing of protein using unmodified gold nanoparticle probes. Chem Commun (Camb) 36:3735–3737
Xue X, Wang F, Liu X (2008) One-step, room temperature, colorimetric detection of mercury (Hg2+) using DNA/nanoparticle conjugates. J Am Chem Soc 130:3244–3245
Tan YN, Su X, Zhu Y, Lee JY (2010) Sensing of transcription factor through controlled-assembly of metal nanoparticles modified with segmented DNA elements. ACS Nano 4:5101–5110
Jung Y, Kim T, Park H, Tom Soh H (2010) Specific colorimetric detection of proteins using bidentate aptamer-conjugated polydiacetylene (PDA) liposomes. Adv Funct Mater 20:3092–3097
Liu J, Mazumdar D, Lu Y (2006) A simple and sensitive “dipstick” test in serum based on lateral flow separation of aptamer-linked nanostructures. Angew Chem Int Ed Engl 45:7955–7959
Su S, Nutiu R, Filipe CD, Li Y, Pelton R (2007) Adsorption and covalent coupling of ATP-binding DNA aptamers onto cellulose. Langmuir 23:1300–1302
Patolsky F, Lichtenstein A, Willner I (2003) Highly sensitive amplified electronic detection of DNA by biocatalyzed precipitation of an insoluble product onto electrodes. Chemistry 9:1137–1145
Radi A (2011) Electrochemical aptamer-based biosensors: recent advances and perspectives. Int J Elect Article ID 863196
Min K, Song KM, Cho M, Chun YS, Shim YB, Ku JK, Ban C (2010) Simultaneous electrochemical detection of both PSMA (+) and PSMA (−) prostate cancer cells using an RNA/peptide dual-aptamer probe. Chem Commun (Camb) 46:5566–5568
Ilgu M, Wang T, Lamm M, Nilsen-Hamilton M (2013) Investigating the malleability of RNA aptamers. Methods. doi:10.1016/j.ymeth.2013.03.016
Http://www.clinicaltrials.gov (2013). Accessed 5 June 13
Ni X, Castanares M, Mukherjee A, Lupold S (2011) Nucleic acid aptamers: clinical applications and promising new horizons. Curr Med Chem 18:4206–4214
Bae O-N (2012) Targeting von Willebrand factor as a novel anti-platelet therapy; application of ARC1779, an Anti-vWF aptamer, against thrombotic risk. Arch Pharm Res 35:1693–1699
Mayer G, Rohrbach F, Pötzsch B, Müller J (2011) Aptamer-based modulation of blood coagulation. Hamostaseologie 4:258–263
Cerchia L, Esposito C, Camorani S, Catuogno S, Franciscis V (2011) Coupling aptamers to short interfering RNAs as therapeutics. Pharmaceuticals 4:1434–1449
Zhou J, Bobbin M, Burnett J, Rossi J (2012) Current progress of RNA aptamer-based therapeutics. Front Genet 3:234
Esposito C, Catuogno S, Franciscis V, Cerchia L (2011) New insight into clinical development of nucleic acid aptamers. Discov Med 61:487–496
Acknowledgments
Funding from Department of Science and Technology (DST), Govt. of India, and research facility provided by Indian Institute of Science Education and Research (IISER), Pune is gratefully acknowledged.
Disclosure
Jayeeta Banerjee declares no conflict of interests. Marit Nilsen-Hamilton declares ownership of Molecular Express Inc., Ames, which is exploring applications of aptamers for combating specific diseases including cancer.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Banerjee, J., Nilsen-Hamilton, M. Aptamers: multifunctional molecules for biomedical research. J Mol Med 91, 1333–1342 (2013). https://doi.org/10.1007/s00109-013-1085-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-013-1085-2