Abstract
Estimation of time since death is one of the challenges in forensic science. There are many approaches to estimate the postmortem interval, including both physical and thanatochemistry methods. Decomposition is triggered by a process called autolysis, which induces destructive changes in the cell leading to cell death. Based on the process of cell death signaling, this study analyzed the early postmortem interval (2–8 h since death) using the study of the mRNA expression of Fas Ligand (FasL) and phosphatase and tensin homologue deleted on chromosome 10 (PTEN) by Quantitative-PCR. Results of the study indicate a time-dependent increase in the mRNA levels of both proteins up until 6 h after death. Using a regression analysis in these first 6 h, a positive linear correlation was found between the mRNA expression of these proteins and the time since death. Since PTEN and FasL are implicated in signaling pathways, both proteins are potential candidates to analyze the time since death in time intervals of 6 h or less. Further research is needed to find additional cell death markers and expand the time period for time since death estimation.
Similar content being viewed by others
Introduction
In legal medicine, the accurate determination of the time of death is important due to its role in elucidating possible criminal acts and determining appropriate civil repercussions.
The determination of the postmortem interval can include estimates spanning 1 day to many years after death by using a multidisciplinary approach. The analysis of the postmortem interval is based on the different changes that a corpse suffers after death including: physical processes (body cooling and hypostasis); metabolic processes (supravital reactions); autolysis (loss of selective membrane permeability, diffusion); physicochemical processes (rigor mortis); bacterial processes (putrefaction) [1–5]; and the effect of insect activity [6, 7]. According to these processes, the methods of estimating the time since death can be quantitative, like the measurement of body cooling or potassium in vitreous humor [1], or qualitative, such as subjective descriptions of gastric contents [8, 9].
Numerous techniques have been proposed in the last 60 years for the determination of the time since death by chemical means [10], leading to the emersion of a field called “thanatochemistry”. Within this field, all analyses included in the postmortem chemical changes can be quantified, providing a mathematical description of the postmortem alterations by taking into account influencing factors, such as temperature. Scientific efforts are now seeking to improve these measurements, finding those with the precision and accuracy necessary for the calculation of the postmortem interval. These methods include the measure of volatile fatty acids in the soil solution, which may aid in determining a long time since death interval [11, 12], as well as the analysis of amino acids, neurotransmitters, and decompositional by-products [13].
In spite of past thanatochemistry studies, few reports have analyzed the expression of RNA in the body. After death, RNA is degraded by ribonucleases present in the cell and/or originating from bacteria or other environmental contamination [14]. The diverse studies of RNA postmortem have shown that, depending on the tissue, RNA can be either more or less stabile, demonstrating rapid degradation in the pancreas and liver due to increase in ribonucleases [15], with the highest stability found in human and rat brain tissue [16–19]. While these studies found a correlation between RNA degradation and the postmortem interval, the majority of them were developed by means of using RNA as a possible diagnostic tool.
Decomposition begins ~4 min after death with a process called autolysis. During decomposition, the cells will be progressively destroyed. As a consequence, there is a release and damage of cellular components and metabolites. Few reports have analyzed changes associated with the time since death within the first hours of death [20–23].
The aim of this research was to study the early postmortem interval, between 2 and 8 h, using the analysis of the mRNA expression of Fas Ligand (FasL) and phosphatase and tensin homologue deleted on chromosome 10 (PTEN) by Quantitative-PCR, and to assess the reliability of this method for mRNA in specimens from dead bodies.
Materials and methods
Animal protocol
Four adult male Wistar rats (250–300 g, aged 3 months) were housed individually in metabolism cages under standard lighting (light on 01:00–13:00 hours), temperature (22–24 °C), and relative humidity (55–65 %) conditions. All rats were housed and handled daily under laboratory conditions for at least 15 days before the study. Water and food (Harlan 2014) were available ad libitum. The appropriate animal care and use committees approved all experimental protocols, and the guidelines of the International Public Health Service Guide for the Care and Use of Laboratory Animals were followed. The experiment was developed in the same conditions of light, temperature, and relative humidity described above. Rats were placed in supine position in the bench and killed under anesthesia with intraperitoneal injection of Rompun R (Bayer, Leverkusen, Germany) and left at room temperature between 0 and 8 h postmortem. Immediately after death, as a time 0 or control, 20 mg of gastrocnemius muscle were biopsied from each rat. This tissue is mainly used in rigor mortis studies [24], and it is easy to access and take several samples from the body. From 2 until 8 h after death, 20 mg of this muscle were collected bi-hourly.
RNA isolation and quantification
The samples were homogenized and total RNA was extracted using the GenElute Mammalian Total RNA Miniprep Kit (Sigma, St. Louis, MO, USA), according to the manufacturer’s protocol.
RNA quantification and quality was assessed using NanoDrop 2000c (Thermo Scientific, Wilmington, DE, USA). The RNA levels were measured as a concentration (ng/μl).
cDNA synthesis
RNA was subjected to reverse transcription using the High-Capacity cDNA reverse transcription Kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s protocol. Briefly, 2× RT master mix was prepared: 2 μl 10× RT buffer, 0.8 μl 25× dNTP mix (100 mM), 2 μl 10× RT Random Primers, 1 μl Multiscribe Reverse Transcriptase, and 4.2 μl RNase free water. Next, 10 μl of the RNA sample was added to the master mix. Thermocycling conditions were as follows: 25 °C 10 min, 37 °C 120 min, 85 °C 5 min.
Real-time PCR
Quantitative analysis of mRNA levels of FasL (implicated in cell death signaling and inflammation) and PTEN (inhibitor of PI3K/Akt pathway, which promotes cell proliferation) as well as housekeeper gene, GAPDH (glyceraldehide 3-phosphate dehydrogenase), were assessed by real-time PCR, performed in a HT7900 Real-time PCR system (Applied Biosystems), using SYBRGreen PCR Core Reagents (Applied Biosystems). Thermocycling conditions were as follows: denaturation for 15 s at 95 °C and annealing/extension for 1 min at 60 °C, during 40 cycles. One additional step, a melting curve, was added to distinguish specific from nonspecific products and primer dimers. The melting curve was constructed by increasing the temperature from 60 to 95 °C with a temperature transition rate of 0.2 °C/min. Each sample was tested in triplicate, and analyses of relative gene expression data were done using the 2ΔCT method.
Statistical analysis
Plots of mRNA levels were performed using Sigma Plot software (Systat Software, San Jose, CA, USA). Regression analyses were developed using Microsoft Excel 2011 for Mac (Microsoft, 2010).
Results
Recovery of total RNA
A total of 20 muscle samples were obtained in an 8-h period. RNA quantification showed variability not only between time periods but also even between subjects (Table 1). The first time period, (0 or control), has the highest and most homogenized concentration amongst subjects, (12.1–5.1 ng/μl), as well as the last time period, (8 h), with concentrations between 14.3 and 3.4 ng/μl. The lowest concentration was found in the samples collected 6 h after death (2.6–1.4 ng/μl). However, in the conversion to cDNA, we adjusted all the concentrations to 10 ng in order to have the same amount of starting RNA.
Real-time PCR for mRNA of cell death proteins: PTEN and FasL
After the reverse transcription, real-time PCR was performed to analyze the expression levels of PTEN and FasL, using 0 h as control. In addition, we used the reference gene (GAPDH) to normalize possible variations during RNA isolation, retro-transcription, or Q-PCR efficiencies, as the expression levels of a reference gene remain consistent under experimental conditions or different tissues.
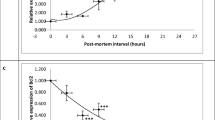
The results were similar in both genes. PTEN showed a time-dependent increase in mRNA levels, starting 2 h after death. However, 8 h after death, mRNA levels suddenly decreased (Fig. 1a). We found the same pattern in mRNA levels of FasL: there was a time-dependent increase until 6 h after death and a marked decrease at 8 h after death (Fig. 1b). This rapid decrease in the mRNA levels is likely due to degradation of RNA as a consequence of the progress of the autolysis process.
Correlation between mRNA levels of PTEN and FasL and early time since death
The results of mRNA levels correlated with the early postmortem interval. However, because of the sudden decrease at 8 h, the correlations were performed only between 0 and 6 h after death (Fig. 2).
Correlation between expression levels of PTEN (a) and FasL (b) and time since death (TSD). Both mRNA expression levels of these proteins showed a strong positive linear correlation with the TSD from 2 to 6 h. The strength of correlation is marked by “r” and the regression plot: y TSD, x mRNA expression
mRNA levels of PTEN showed a strong positive linear correlation with the early time since death (r = 0.98; p = 0.01; with the regression formula TSD = 0.04mRNA levels + 0.25). The same high positive linear correlation was found in mRNA levels of FasL (r = 0.94; p = 0.05; with the regression formula TSD = 0.01mRNA levels + 1.02). These results verified our initial hypothesis, since FasL and PTEN are implicated in cell death signaling pathways.
Discussion
The findings from this research provide a quantitative tool for estimating the early postmortem interval until 6 h after death. We used the analysis of the expression of two genes implicated in living organisms on cell death and inflammation: PTEN and FasL (Fig. 3).
Signaling pathways of cell death. PTEN is implicated in dephosphorylation of PIP3, which is the product of PI3K. PIP3 activates AKT by its kinase. AKT promotes cell survival keeping in the cytoplasm, in addition to others, FKHRL, implicated in FasL transcription. When PTEN converts PIP3 in PIP2, AKT is neutralized and FKHRL goes to the nucleus and triggers apoptotic signals, like FasL. When FasL binds to its receptor, DISC is formed, activating Caspase-8, which in turns activates the effector Caspase-3. If the signal of Caspase-8 is not enough to induce cell death, the mitochondrial pathway is activated with the regulator Caspase-9, which triggers the activation of Caspase-3. This sequence of events demonstrates the relationship of the expression levels between PTEN and FasL. The decrease in the expression of both proteins 8 h after death could be due to degradation of RNA or the increase of the oxidative stress from the cell death process, which inactivates PTEN
PTEN [25] was identified as a tumor-suppressor gene located in a critical region of chromosome 10, 10q23.3, and was found to be deleted in a wide variety of human cancers and cell lines, including glioblastomas, prostate, breast, and kidney cancer cell lines [26]. The amino acid sequence of PTEN indicated that it resembles two different types of proteins. The PTEN genes encode the catalytic signature motif of proteins tyrosine phosphatases and functions as a dual-specificity phosphatase, but it can also dephosphorylate the lipid signal transduction molecule (PIP3) (phosphatidylinositol 3, 4, 5-trisphosphate) [26, 27]. The N-terminal domain of PTEN also shows extensive homology to the cytoskeletal protein tensin, which plays a role in the maintenance of cellular structure and possibly signal transduction by binding to actin filaments [28]. All these functions have implicated PTEN in the regulation of several different cellular processes: cell growth, apoptosis (programmed cell death), interactions with the extracellular matrix, and cell migration and invasion [25].
The regulatory function of PTEN in cell death was shown in this work by an increase in expression with the time after death. The regulatory pathway starts with the dephosphorylation of phosphatidylinositol 3,4,5 triphosphate (PtdIns(3,4,5)P3) and phosphatidylinositol 3,4 biphosphate (PtdIns(3,4)P2), resulting in PtdIns(4,5)P and PtdIns(4)P. These phospholipids are the direct products of the phosphatidylinositol 3-OH kinase (PI-3 kinase, PI3K), which counteracts PTEN function (Fig. 3, left). PI3K is activated by growth factor stimulation, which is followed by phosphorylation of inositol substrates at position 3, to produce phosphatidylinositol 3,4,5 triphosphate [PtdIns(3,4,5)P3], which in turn activates serine-threonine AKT and AKT kinases, like PDK1 (phosphonositide-dependent kinase 1), which phosphorylates and activates AKT or PKB (protein kinase B). Activated AKT promotes cell survival through multiple pathways [29]. One of the pathways implies the suppression of FasR/FasL-induced apoptosis, by AKT regulation of the activity of the Forkhead Transcription Factor, FKHRL1. In the presence of survival factors, AKT phosphorylates FKHRL1, which keeps it in the cytoplasm. PTEN neutralizes the action of AKT, permitting FKHRL1 to go to the nucleus and trigger apoptotic signals by inducing the expression of cell death genes like FasL (Fig. 3, center) [30, 31].
These studies demonstrate the link between PTEN and FasL, since PTEN is implicated in the regulation of FasL and death signals. In the present study, we saw that the expression of both genes is parallel, having a time-dependent increase until 6 h after death. At 8 h, we found a decrease in their expression. This could be due to a degradation of RNA postmortem, although several studies have demonstrated that RNA could be stable several days after death depending on the tissue [14]. However, another explanation could be the process of death itself. The signaling of FasL is activated by cytotoxic stress and DNA damage (Fig. 3, right) [32]. Binding of FasL with its receptor, Fas triggers the formation of the death-inducing signaling complex (DISC) by recruiting an adaptor molecule FADD (Fas-associating protein with death domain) to the cytoplasmic tail of Fas (C-terminal region). FADD recruits procaspase 8, which is immediately proteolytically processed to its active larger form. Caspase-8 activates Caspase-3, which is the effector caspase to activate the cell death signaling in the nucleus. Sometimes, the Caspase-8 signal is not enough to induce cell death, and for that reason this protein activates Bid (a cytoplasmic protein) to activate the intrinsic-mitochondrial pathway of apoptosis with the release of cytochrome c, which is recruited to form the apoptosome with Apaf and procaspase 9. Activated Caspase-9 triggers the activation of Caspase-3, resulting in cell death [33]. Both cell death and the activation of mitochondrial pathway of apoptosis increase the free radicals generated by the Electron Transport Chain, and this increase in the oxidative stress could lead to the inactivation through oxidation of PTEN [34]. In our study, 8 h after death it is possible that the increase in free radicals induces a deactivation of PTEN that in turn cannot activate FasL, leading to the autolysis process.
The course of decomposition is similar to the process induced when an organ suffers ischemic or anoxic alterations. Several studies have addressed the implication of PTEN and FasL in the ischemia process [34–39]. In fact, in ischemic/reperfused myocardium, the antagonist of PTEN, AKT plays a protector role by reducing the myocardial infarction, PTEN being the cause of the death of myocardial cells [34]. FasL were found unregulated in neuronal cells undergoing apoptosis after ischemia [35]. In contrast, and as a protective intrinsic mechanism, PTEN was phosphorylated and therefore inactivated by brain ischemia [40, 41].
The role of PTEN and FasL in cell death as well as their implication in ischemia makes them potential candidates to analyze the postmortem interval in the first hours of death. However, their decrease after 8 h, likely due to RNA degradation and increase in free radicals, requires more research, not only in these genes but also within additional cell death markers to extend both the time period and accuracy of time since death estimation.
References
Madea B (1993) Time since death extrapolated from vitreous potassium. Forensic Sci Int 59(1):80–82
Madea B, Henssge C, Honig W, Gerbracht A (1989) References for determining the time of death by potassium in vitreous humor. Forensic Sci Int 40(3):231–243
Madea B, Herrmann N, Henbge C (1990) Precision of estimating the time since death by vitreous potassium—comparison of two different equations. Forensic Sci Int 46(3):277–284
Madea B, Kaferstein H, Hermann N, Sticht G (1994) Hypoxanthine in vitreous humor and cerebrospinal fluid–a marker of postmortem interval and prolonged (vital) hypoxia? Remarks also on hypoxanthine in SIDS. Forensic Sci Int 65(1):19–31
Madea B, Kreuser C, Banaschak S (2001) Postmortem biochemical examination of synovial fluid—a preliminary study. Forensic Sci Int 118(1):29–35
Benecke M (2001) A brief history of forensic entomology. Forensic Sci Int 120(1–2):2–14
Amendt J, Krettek R, Zehner R (2004) Forensic entomology. Naturwissenschaften 91(2):51–65. doi:10.1007/s00114-003-0493-5
Henssge C, Madea B (1988) Determination of the time since death. I. Body heat loss and classical signs of death. An integrated approach. Acta Med Leg Soc 38(1):71–89
Henssge C, Madea B, Gallenkemper E (1988) Death time estimation in case work. II. Integration of different methods. Forensic Sci Int 39(1):77–87
Madea B (2005) Is there recent progress in the estimation of the postmortem interval by means of thanatochemistry? Forensic Sci Int 151(2–3):139–149. doi:10.1016/j.forsciint.2005.01.013
Vass AA, Bass WM, Wolt JD, Foss JE, Ammons JT (1992) Time since death determinations of human cadavers using soil solution. J Forensic Sci 37(5):1236–1253
Vass AA (2011) The elusive universal post-mortem interval formula. Forensic Sci Int 204(1–3):34–40. doi:10.1016/j.forsciint.2010.04.052
Vass AA, Barshick SA, Sega G, Caton J, Skeen JT, Love JC, Synstelien JA (2002) Decomposition chemistry of human remains: a new methodology for determining the postmortem interval. J Forensic Sci 47(3):542–553
Bauer M (2007) RNA in forensic science. Forensic Sci Int Genet 1(1):69–74. doi:10.1016/j.fsigen.2006.11.002
Finger JM, Mercer JF, Cotton RG, Danks DM (1987) Stability of protein and mRNA in human postmortem liver—analysis by two-dimensional gel electrophoresis. Clin Chim Acta 170(2–3):209–218
Johnson SA, Morgan DG, Finch CE (1986) Extensive postmortem stability of RNA from rat and human brain. J Neurosci Res 16(1):267–280. doi:10.1002/jnr.490160123
Chung WH, Hung SI (2012) Recent advances in the genetics and immunology of Stevens–Johnson syndrome and toxic epidermal necrosis. J Dermatol Sci 66(3):190–196. doi:10.1016/j.jdermsci.2012.04.002
Bauer M, Gramlich I, Polzin S, Patzelt D (2003) Quantification of mRNA degradation as possible indicator of postmortem interval—a pilot study. Leg Med (Tokyo) 5(4):220–227
Inoue H, Kimura A, Tuji T (2002) Degradation profile of mRNA in a dead rat body: basic semi-quantification study. Forensic Sci Int 130(2–3):127–132
Elmas I, Baslo B, Ertas M, Kaya M (2001) Analysis of gastrocnemius compound muscle action potential in rat after death: significance for the estimation of early postmortem interval. Forensic Sci Int 116(2–3):125–132
Elmas I, Baslo MB, Ertas M, Kaya M (2002) Compound muscle action potential analysis in different death models: significance for the estimation of early postmortem interval. Forensic Sci Int 127(1–2):75–81
Liu L, Shu X, Ren L, Zhou H, Li Y, Liu W, Zhu C, Liu L (2007) Determination of the early time of death by computerized image analysis of DNA degradation: which is the best quantitative indicator of DNA degradation? J Huazhong Univ Sci TechnoloMed Sci 27(4):362–366. doi:10.1007/s11596-007-0404-7
McDowall KL, Lenihan DV, Busuttil A, Glasby MA (1998) The use of absolute refractory period in the estimation of early postmortem interval. Forensic Sci Int 91(3):163–170
Huang H, Yan Y, Zuo Z, Yang L, Li B, Song Y, Liao L (2010) Determination of adenosine phosphates in rat gastrocnemius at various postmortem intervals using high performance liquid chromatography. J Forensic Sci 55(5):1362–1366. doi:10.1111/j.1556-4029.2010.01450.x
Tamura M, Gu J, Tran H, Yamada KM (1999) PTEN gene and integrin signaling in cancer. J Natl Cancer Inst 91(21):1820–1828
Li DM, Sun H (1997) TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res 57(11):2124–2129
Maehama T, Dixon JE (1998) The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 273(22):13375–13378
Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV (1997) Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet 15(4):356–362. doi:10.1038/ng0497-356
Dahia PL (2000) PTEN, a unique tumor suppressor gene. Endocr Relat Cancer 7(2):115–129
Rohn JL, Hueber AO, McCarthy NJ, Lyon D, Navarro P, Burgering BM, Evan GI (1998) The opposing roles of the Akt and c-Myc signalling pathways in survival from CD95-mediated apoptosis. Oncogene 17(22):2811–2818. doi:10.1038/sj.onc.1202393
Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96(6):857–868
Pinkoski MJ, Green DR (1999) Fas ligand, death gene. Cell Death Differ 6(12):1174–1181. doi:10.1038/sj.cdd.4400611
Kavurma MM, Khachigian LM (2003) Signaling and transcriptional control of Fas ligand gene expression. Cell Death Differ 10(1):36–44. doi:10.1038/sj.cdd.4401179
Mocanu MM, Yellon DM (2007) PTEN, the Achilles’ heel of myocardial ischaemia/reperfusion injury? Br J Pharmacol 150(7):833–838. doi:10.1038/sj.bjp.0707155
Martin-Villalba A, Herr I, Jeremias I, Hahne M, Brandt R, Vogel J, Schenkel J, Herdegen T, Debatin KM (1999) CD95 ligand (Fas-L/APO-1L) and tumor necrosis factor-related apoptosis-inducing ligand mediate ischemia-induced apoptosis in neurons. J Neurosci 19(10):3809–3817
Zhao ZQ, Corvera JS, Halkos ME, Kerendi F, Wang NP, Guyton RA, Vinten-Johansen J (2003) Inhibition of myocardial injury by ischemic postconditioning during reperfusion: comparison with ischemic preconditioning. Am J Physiol Heart Circ Physiol 285(2):H579–H588. doi:10.1152/ajpheart.01064.2002
Zhu M, Feng J, Lucchinetti E, Fischer G, Xu L, Pedrazzini T, Schaub MC, Zaugg M (2006) Ischemic postconditioning protects remodeled myocardium via the PI3K-PKB/Akt reperfusion injury salvage kinase pathway. Cardiovasc Res 72(1):152–162. doi:10.1016/j.cardiores.2006.06.027
Tong H, Chen W, Steenbergen C, Murphy E (2000) Ischemic preconditioning activates phosphatidylinositol-3-kinase upstream of protein kinase C. Circ Res 87(4):309–315
Mocanu MM, Yellon DM (2003) p53 down-regulation: a new molecular mechanism involved in ischaemic preconditioning. FEBS Lett 555(2):302–306
Omori N, Jin G, Li F, Zhang WR, Wang SJ, Hamakawa Y, Nagano I, Manabe Y, Shoji M, Abe K (2002) Enhanced phosphorylation of PTEN in rat brain after transient middle cerebral artery occlusion. Brain Res 954(2):317–322
Choi JS, Park HJ, Kim HY, Kim SY, Lee JE, Choi YS, Chun MH, Chung JW, Lee MY (2005) Phosphorylation of PTEN and Akt in astrocytes of the rat hippocampus following transient forebrain ischemia. Cell Tissue Res 319(3):359–366. doi:10.1007/s00441-004-1033-0
Acknowledgments
Sara C. Zapico is supported by a Peter Buck Postdoctoral Fellowship from the Smithsonian Institution. The authors gratefully acknowledge Keitlyn Alcántara-Russell for the language revision of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
C. Zapico, S., Menéndez, S.T. & Núñez, P. Cell death proteins as markers of early postmortem interval. Cell. Mol. Life Sci. 71, 2957–2962 (2014). https://doi.org/10.1007/s00018-013-1531-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-013-1531-x