Abstract
Chronic rhinosinusitis with nasal polyps is considered a subgroup of chronic rhinosinusitis and a significant health problem, but the pathogenesis remains unclear to date. Therefore, we investigated the stemness to determine the role of stem cells in nasal polyps, with additional analysis of the neuronal differentiation potential of nasal polyp cells. We determined gene and protein expression profiles of stem cells in nasal polyp tissues, using whole genome microarray, quantitative real-time PCR (qPCR), immunohistochemistry, and flow cytometry. To evaluate the neuronal differentiation potential of nasal polyp cells, we used an efficient xenogeneic co-culture model with unsliced adult rat brain biopsies, followed by qPCR, immunohistochemistry, and growth factor antibody arrays. During gene expression analysis and immunohistochemistry, we were able to detect different stem cell markers, like Oct-4, Sox2, Klf4, c-Myc, ABCG2, Nanog, CD133, and Nestin, which confirmed the existence of stem cell like cells within nasal polyps. In addition, co-culture experiments give evidence for a guided differentiation into the neuronal lineage by overexpression of Nestin, Neurofilament, and GM-CSF. Our study demonstrated the expression of stem cell-related markers in nasal polyps. Furthermore, we characterized, for the first time, the stemness and neuronal differentiation potential of nasal polyp cells. These results gave new insights into the pathogenesis of nasal polyps and its therapeutic effectiveness could represent a promising strategy in the future.
Similar content being viewed by others
Introduction
Chronic rhinosinusitis with nasal polyps (CRSwNP) is considered a subgroup of chronic rhinosinusitis, a chronic inflammatory condition of the nasal and paranasal sinuses, and is characterized by grape-like structures in the upper nasal cavity. Typical histological features of nasal polyps are dense inflammatory infiltrates, loose fibrous connective tissue with substantial tissue edema, and a thickened basement membrane covered mostly by respiratory pseudostratified epithelium with ciliated cells and goblet cells (Fokkens et al. 2012). The nasal epithelium is constantly encountered to environmental factors like allergens, bacteria, or viruses, and is the first line of defense against these factors, but it serves not only as physical barrier. Nasal epithelial cells are able to detect and respond to environmental signals by producing cytokines which mediate the recruitment of immunocompetent cells and create a micro-environment for these cells (Vroling et al. 2008).
Under chronic inflammatory conditions, the epithelium must regenerate to restore its defensive functions (Watelet et al. 2006). This regenerative process may involve multilineage stem/progenitor cells with self-renewal and proliferation capacities (Alison and Islam 2009). Adult stem cells are undifferentiated cells found in different tissues and organs (Alison and Islam 2009). They have common attributes which afford them self-renewal, survival, and conservation of genomic integrity, and are located in specialized micro-environments (Alison and Islam 2009; Kim et al. 2009; Randell 2006). In the development and progression of malignant tumors, cells of this type could drive continued expansion of malignant cells (Jordan et al. 2006), but the underlying molecular mechanisms of the benign cell proliferation and the chronic recurrent nature of nasal polyps are still unknown.
The previous studies about stem cells in nasal polyps dealt with the identification and characterization of specific stem cell populations. In detail, researchers linked the pathogenesis of nasal polyps to the presence and differentiation of mesenchymal stem cells (MSC) (Cho et al. 2014, 2015; Pezato et al. 2014), pathological changes in the epithelium (de Borja Callejas et al. 2014; Li et al. 2014; Wiszniewski et al. 2006; Yu et al. 2014) or to single stem cell markers like stem cell factor, Nestin, and BMI-1 (Kim et al. 1997, 2009; Kowalski et al. 2005). The overall stemness and for example neuronal differentiation potential of nasal polyps are still unknown. The aim of this study was to clarify the expression of further stem cell markers and to characterize the stemness in an effort to determine the role of stem cells in nasal polyps. In addition, we evaluated the neuronal differentiation potential of nasal polyp cells from chronic rhinosinusitis patients in an efficient xenogeneic co-culture model.
Materials and Methods
Ethics Statements
The study was approved by the local ethics committee of the University of Schleswig–Holstein, Campus Lübeck and conducted in accordance with the ethical principles for medical research formulated in the WMA Declaration of Helsinki. All participants were treated surgically at the Department of Otorhinolaryngology, University Hospital Schleswig–Holstein, Campus Lübeck, and have given their written informed consent.
The isolation of brain biopsies from Sprague–Dawley rats was approved by the animal protection committee of the University Lübeck.
Patient Specimens
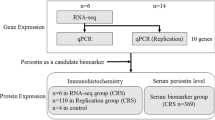
Nasal polyp tissue and associated inferior turbinate tissue, as internal control, were harvested from 26 patients (21 males and 5 females, mean age: 51.18 ± 18.26) who underwent functional endoscopic sinus surgery. Fresh tissue samples were flash frozen in liquid nitrogen immediately after resection, stored at −80 °C before RNA and protein extraction, and additionally embedded for cryostat sections and used for dissociation of nasal polyps. For microarray analysis, nasal polyp tissue and inferior turbinate tissue of 8 patients (7 males and 1 female, mean age: 53.87 ± 13.34) were representatively analyzed.
All patients had a history of sinusitis of more than 3 months and did not respond to conservative therapy. Patients were skin tested for pollens, molds, dust mites, and pets using standardized extracts (Allergopharma Joachim Ganzer KG, Reinbek, Germany) within a time frame of 4 weeks before surgery. Eosinophilic CRSwNP was determined by histopathologic examination and patients with mucoviscidosis or neutrophilic nasal polyps were not included in this study. All patients had been free of steroid medication for at least 4 weeks before surgery and had no history of atopy, bronchial asthma, or salicylate intolerance/aspirin-exacerbated respiratory disease.
Microarrays
To identify the basal gene expression profiles of nasal polyps and inferior turbinates, we performed Agilent Whole Human Genome Microarrays (4 × 44 K, Miltenyi Biotec, Bergisch Gladbach, Germany).
Frozen tissue samples were shipped on dry ice to Miltenyi Biotec (Bergisch Gladbach, Germany) for microarray analysis. RNAs were isolated using standard RNA extraction protocols (Trizol) and were quality-checked via the Agilent 2100 Bioanalyzer platform (Agilent Technologies). The RNA integrity number (RIN) value was calculated and RNA with an RIN number >6 was used (Fleige and Pfaffl 2006). Agilent whole human genome microarrays (4 × 44 K) were performed following the manufacturer’s protocols. The Rosetta Resolver® gene expression data analysis system (Rosetta Biosoftware) was used to compare two single intensity profiles in a ratio experiment (inferior turbinate versus nasal polyp). These experiments adhere to the Minimal Information About A Microarray Experiment guidelines.
Quantitative Real-Time PCR
Quantitative real-time PCR (qPCR) was performed to confirm microarray results and to analyze the transcription of markers for stemness, proliferation, and differentiation to all three germ layers, using TaqMan® Gene Expression Assays (Applied Biosystems, Foster City, CA, USA; Table 1). The transcriptional activity of studied genes was analyzed using a LightCycler 1.5 (Roche, Mannheim, Germany). One microgram of each RNA sample was synthesized to cDNA using the RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, St. Leon-Rot, Germany) according to the manufacturer’s instructions. qPCR reaction mixture consisted of 10 µl TaqMan® Gene Expression Master Mix (Applied Biosystems, Foster City, CA, USA), 1 µl TaqMan® Gene Expression Assay (Applied Biosystems, Foster City, CA, USA), and 5 ng of cDNA. Detailed program information was published before (Könnecke et al. 2014). To ensure that the reference gene was stably expressed in nasal polyps and inferior turbinates, we tested several housekeeping genes such as β-actin, GAPDH, or HPRT1 according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments Guidelines. Beta-actin was invariantly expressed under experimental conditions, thus all selected gene mRNA levels in patients were measured and normalized to β-actin. The 2−∆∆Ct method (Livak and Schmittgen 2001) was used to analyze the qPCR data. In addition, agarose gel electrophoresis was applied on generated PCR-products in the course of the qualitative characterization of mRNA-products in nasal polyp tissue.
Immunohistochemistry
Frozen tissue sections were air dried and fixed in ice-cold (−20 °C) acetone for 10 min. Immunostaining was performed using the Labeled Streptavidin Biotin method. Sections were incubated for 15 min with 3% H2O2, rinsed three times in Tris-buffered saline (TBS) and incubated overnight with rabbit monoclonal antibody Sox2 (1:100, GeneTex, USA). After washing in TBS, sections were incubated with polylink biotinylated antibody for 20 min, followed by peroxidase labeled avidin-biotin-complex for 20 min (Dako, Denmark). Visualization was achieved by a final incubation with 3-amino-9-ethylcarbazol-peroxidase substrate chromogen. Sections were counterstained with Mayer’s hematoxylin and analyzed with an Axiovert 200 M microscope (Carl Zeiss AG, Oberkochen, Germany).
Immunofluorescence
Frozen tissue sections were air dried and fixed in 4% paraformaldehyd in phosphate-buffered saline (PBS; Gibco, Life Technologies, Carlsbad, CA, USA) containing 0.1% TritonX. Non-specific binding was blocked by incubation with 20% normal serum (Vector Laboratories, CA, USA) diluted in PBS for at least 20 min at room temperature. Tissue sections were incubated with primary antibody against α-SMA (1:1000, mouse; Dako, Denmark), Cytokeratin 18 (CK18, 1:200, mouse; Santa Cruz, USA), Ki67 (1:500, rabbit; Abcam PLC, USA), Nestin (1:100, mouse; Millipore, USA), Neurofilament-mix (NF, 1:500, rabbit; GeneTex, USA), and Oct-4A (20 µg/ml, mouse; Santa Cruz, USA) diluted in TBS with 1% Tween20 containing 0.1% bovine serum albumin in a humid chamber over night at 4 °C. After rinsing three times with PBS, tissue sections were incubated with fluorescence labeled secondary antibody in a humid chamber for 60 min at 37 °C. After washing three times in PBS, tissue sections were covered with Vectashield mounting medium (Vector Laboratories, CA, USA) and analyzed with a laser scanning microscope (LSM 710; Carl Zeiss AG, Germany).
Preparation of Single Cell Suspension
Tissue samples were washed in PBS, carefully minced into small pieces with a scalpel blade, and digested by enzymatic treatment in an enzyme mixture solution of collagenase type II (31.5 mg/ml; Gibco, Life Technologies, Carlsbad, CA, USA), hyaluronidase (3.99 mg/ml; Sigma, Munich, Germany) and dispase (33.4 mg/ml; Sigma, Munich, Germany). One milliliter of each enzyme was added to 15 ml sterile serum free high-glucose Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Life Technologies, Carlsbad, CA, USA) containing 1% l-glutamine and supplemented with 10% fetal bovine serum (FBS; Gibco, Life Technologies, Carlsbad, CA, USA), 2.5% 25 mM HEPES buffer, 1% sodium pyruvate, 1% nonessential amino acids and 5–10% of antibiotic/antimycotic solution (Gibco, Life Technologies, Carlsbad, CA, USA) under shaking at 37 °C for 120 min. After enzymatic treatment, cells were washed and centrifuged. Then, the dissociated cells were washed in PBS, and filtered through a 70 and a 40 µm-nylon cell strainer (Falcon, Becton Dickinson Labware, Heidelberg, Germany) to generate a single cell suspension. The resulting cell suspensions were cultured and then used for flow cytometry and co-culture experiments.
Flow Cytometry
Forty-eight hours after cell isolation, cultured cells were washed in PBS and detached. To remove cell clumps, cells were passed through a 40 µm-nylon cell strainer. Cells were counted and dead cells were excluded by using the trypan blue exclusion assay. 2 × 106 cells were suspended in 50 µl buffer of 2% FBS (Gibco, Life Technologies, Carlsbad, CA, USA) in PBS and then incubated in the dark at 4 °C for 15 min with specific antibody concentrations of 2 µl/50 µl. A surface antigen staining with the lineage cocktail 1 FITC including CD3, CD14, CD16, CD19, CD20, and CD56 (Becton Dickinson, Heidelberg, Germany) was performed for excluding Oct-4-positive lymphocytes, monocytes, eosinophils, and neutrophils. Then, cells were washed by adding the buffer and centrifuged at 300g for 5 min. Supernatant was poured off and cell pellets were suspended again in a 50 µl buffer. Afterwards, for intracellular staining of Oct-4 (rat, R&D, Minneapolis, USA), cells were treated with 50 µl 0.1% Saponin. The percentage of positive cells was measured from a cut-off set determined using the isotype-matched non-specific control antibodies APC IgG2b, PE IgG2b, APC IgG2b, (all R&D, Minneapolis, USA), and FITC IgG2b (Becton Dickinson, Heidelberg, Germany) as internal controls. The levels detected by these isotype control antibodies were used as a basal background which had to be subtracted from each result. 1 × 106 events were analyzed by flow cytometry on a FACSCanto (Becton Dickinson) equipped with the FACS DIVA software. Dead cells were excluded by staining the cells with Propidium Iodide–PE (Becton Dickinson, Heidelberg, Germany).
Co-culture Experiments
Fresh nasal polyp tissues were separated into single cells and cultured in DMEM containing 10% FBS and 1% antibiotics/antifungal agent. Two days prior co-culture experiments, cells were detached and seeded in DMEM containing 10% FBS on 6-well culture plates (18,000 cells/well). Rat brain biopsies were obtained from 8-week-old male rats. For indirect co-culture experiments, one 0.027 cm³ piece of cerebrum and cerebellum each was separated from nasal polyp cells by tissue culture inserts (0.4 µm pore size, Greiner Bio-One). Within this system, growth factors and cytokines from brain tissue and NP cells could pass the porous membrane of the insert, but no cells could cross. Nasal polyp cells cultured without the addition of rat brain biopsies serve as controls (Petschnik et al. 2011).
Growth Factor Antibody Array
To analyze the secretion of growth factors during the co-culturing of human nasal polyp cells with rat brain biopsies, a human growth factor antibody array (Growth Factor Human Membrane Antibody Array, ab134002, Abcam PLC, Cambridge, MA, USA) was performed. Media supernatants of co-cultures (with and without brain) from independently elaborated experiments were brought onto a membrane spotted with antibodies against 41 different growth factors. The procedure was done according to the manufacturer’s instructions. Spots were detected by chemiluminescence on the Fusion FX7 (Vilber Lourmat, Torcy, France). Culture medium without cells and brain and culture medium without cells but with brain were used to assess baseline signal response.
Statistical Analysis
For statistical analysis and graphs, Prism software (GraphPad, San Diego, USA) was used. Experiments were performed in triplicates. Means and standard deviations were compared using Wilcoxon matched-pairs signed rank test. P values ≤0.05 were considered to be statistically significant.
Results
Transcriptional Analysis of Stem Cell Markers in Nasal Polyps
Based on determined genome wide gene expression profiles of nasal polyps and inferior turbinates, the relative gene expression of stem cell-related markers were analyzed. In nasal polyps, many of the traditional stem cell genes and stem cell-associated genes were differentially expressed between the analyzed patients (Fig. 1). To identify special subsets of stem cell markers, we arranged these markers concerning their origin (Fig. 2). However, no clear profile of a specific type of stem cell marker could be detected. The traditional stem cell markers like Oct-4, Sox2, Klf4, Nanog, ABCG2, CD133, Nestin, and c-Myc were differentially expressed (0.55-fold to 1.8-fold). To confirm the data obtained by microarray analysis, we studied the mRNA expression of stem cell markers using qPCR. Median expression of stem cell markers did not significantly differ between nasal polyps and inferior turbinates (0.70–1.22-fold; Fig. 3), except Sox2 was slightly up-regulated (1.64-fold). However, the expression of specific stem cell markers draws conclusions about the existence of stem cells in nasal polyp tissue, even if the expression did not differ compared to turbinate tissue. Further experiments oriented toward the identification of neuronal stem cell within nasal polyps and their neuronal differentiation capability.
Nasal Polyp Cells Demonstrate Spontaneous Multilineage Differentiation Potential
To characterize the stemness and multilineage differentiation potential of nasal polyp cells, we investigated different stem cell markers and markers of the three germ layers on the mRNA and protein level. mRNA expression analysis showed that specific stem cell markers, such as Nestin, Nanog, ABCG2, CD9, c-Myc, Oct-4, and Sox2, could be detected in nasal polyp cells (Fig. 4).
Protein expression of Nestin and Sox2 was confirmed by immunohistochemistry (Figs. 5, 6a, 7). Flow cytometric investigations analyzing single cell suspensions revealed an average of 1.7% of Oct-4 A expressing cells in Polyposis nasi, which were shown to be negative for lineage markers CD3, CD16, CD19, CD20, and CD56 (Fig. 6b). To characterize the proliferation activity of nasal polyp cells, protein expression of the proliferation marker Ki67 was analyzed and revealed proliferating cells in the epithelium of nasal polyps (Fig. 7). In addition, marker proteins of all three germ layers were detectable. In regard to ectodermal differentiation markers, NF and CK18 were enriched on both the mRNA and protein level (Figs. 4, 7). S100ß-mRNA, a marker for Schwann cell differentiation, could be also detected in nasal polyps (Fig. 4). αSMA represents a mesodermal differentiation marker and was discovered on the mRNA and protein level (Figs. 4, 7). Additional mesodermal markers such as MEF2D (myogenic), PPARγ (adipogenic), and SPP1 (osteogenic) were detected at the mRNA level (Fig. 4). Moreover, the endodermal differentiation marker υWF was also detected by mRNA expression (Fig. 4).
Identification of Oct-4 A expressing cells in nasal polyps of chronic rhinosinusitis patients. Immunohistochemical (a) and flow cytometric investigations (b) revealed an average of 1.7% of Oct-4 A expressing cells in nasal polyps, which were shown to be negative for lineage markers CD3, CD16, CD19, CD20, and CD56. This figure is a representative example of several experiments
Co-culture of Nasal Polyps with Rat Brain Biopsies
The co-culture system is suitable for testing neuronal differentiation capability of numerous types of stem cells (Petschnik et al. 2011). We performed qPCR and immunohistological analysis to evaluate the influence of co-cultured rat brain biopsies on the neuronal differentiation capability of nasal polyp cells. In nasal polyp cell populations, only a few cells were positive for Nestin (Fig. 9f). After co-cultivation with rat brain biopsies for a period of 2 days, the amount of Nestin-expressing cells slightly increased (Fig. 9e). This increase could also be measured on the mRNA level. The transcriptional activity of Nestin was 3.2 times higher after co-cultivation (Fig. 8). In the control approach, NF was only low expressed by a small amount of cells (Fig. 9d). Co-cultivation led to an increase of NF-positive cells and a change in the morphological appearance to an elongated and axon-like cell type (Fig. 9c). Consistent with these results, the relative expression of NF mRNA strongly increased (Fig. 8). Alpha SMA served as an indicator for mesodermal differentiation, but the protein expression and transcriptional activity remained unchanged (Figs. 8, 9a, b). In addition, it could be shown that ABCG2 (2.14-fold), another neuronal stem cell marker, was also up-regulated on the mRNA level.
Determination of Secreted Growth Factors in Co-culture of Nasal Polyps
To identify growth factors that may be involved in the neuronal differentiation of nasal polyp cells during co-culturing, we performed human growth factor antibody arrays to test for 41 growth factors (Fig. 10). We detected that nasal polyp cells themselves strongly expressed insulin-like growth factor binding protein 2 (IGFBP-2) and IGFBP-6. Rat brain biopsies separately highly express basic fibroblast growth factor (bFGF). The co-culturing of nasal polyp cells with rat brain biopsies led to a 7.45-fold higher secretion of granulocyte macrophage colony-stimulating factor (GM-CSF) compared to the single approaches. FBS was used in the culture medium and that is the reason why several growth factors were already present at low levels in the untreated supernatants.
Discussion
Stem cells are engaged in homeostasis and tissue repair on different mucosal surfaces (Yu et al. 2012). During the past years, many studies have introduced a variety of stem cell markers (Alison and Islam 2009) identifying special subgroups, correlating diverse functions to marker patterns. Stem cells have an essential role in the development of complex multicellular organisms, but their unique properties, especially the attribute of self-renewal, make them highly relevant to benign repair mechanisms as well as malignancy and oncogenesis (Pardal et al. 2003). Under normal conditions, the airway epithelium is slowly renewed after damage, but it can also proliferate notably to repair an injury (Ayers and Jeffery 1988). Abnormal increased self-renewal combined with intrinsic growth potential of stem cells may result in a malignant phenotype (Jordan et al. 2006). The recurrent nature, the uncontrolled benign cell proliferation of nasal polyps, arising from inflammatory micro-environment parallels steps of stem cell development described in malignancies.
Nasal stem cells should be expected in the nasal epithelium, because it is constantly encountered to environmental factors like allergens, bacteria, or viruses, and is the initial site of injury from these factors (Vroling et al. 2008; Yu et al. 2012). Actually, we were able to identify different stem cell markers which have been already linked to various cell types like induced pluripotent stem cells, hematopoietic stem cells, MSC, or neural stem cells, which confirmed the existence of stem cell like cells in nasal polyps. A recently published study showed that the turbinate mucosa represents different mechanical properties between middle and inferior turbinate of chronic rhinosinusitis patients with nasal polyps (Pezato et al. 2016b). The authors came to the conclusion to avoid using the inferior turbinate as control for nasal polyposis and suggested to use the middle meatus mucosa from healthy nasal mucosa. This is a good point and should be considered in the future. However, for our transcriptional analysis, we used the inferior turbinate of the same patient, because this represents a good internal control due to the fact that it is not affected by nasal polyp growth and shares the same chronic inflammatory conditions as nasal polyps.
Oct-4, a Pou domain-containing transcription factor, has become a proxy for stemness in many different studies of adult stem cells and is an essential mediator of the embryonic stem cell state (Campbell et al. 2007; Lengner et al. 2008). Oct-4 expression has been demonstrated at different locations in the human body like bone marrow, hair follicles, breast, liver, or neural stem cell (Gupta and Rosenberg 2008; Jiang et al. 2002; Reyes et al. 2002; Yu et al. 2006) and now also in nasal polyp cells. Besides Oct-4, Sox2 is one of the transcription factors which is essential for the pluripotent cell development and maintenance of undifferentiated embryonic stem cells (Avilion et al. 2003; Nichols et al. 1998) and was found in the epithelium of nasal polyps. Nestin has been reported not only as an adult stem cell marker (Wiese et al. 2004) but also as a marker for neuroectodermal progenitor cells (Yaworsky and Kappen 1999), and in line with our results, Kim et al. (2009) found Nestin-positive cells in the epithelial basal cell layer of nasal polyps. To figure out if the expression of stem cell-associated genes and proteins in nasal polyps is accompanied with a certain differentiation potential, we determined the cells capability to differentiate along the neuronal lineage. Their prominent Nestin expression let assumes a predisposition for neuroectodermal differentiation. The co-culture approach with rat brain tissue showed a transcriptional increase of Nestin expression, but only a slight increase of Nestin-positive cells. In conjunction with a strong up-regulation of NF, an increase in neuronal differentiated cells was suggested. Relating to the development of neurons (Carden et al. 1987), NF expression appears with the beginning of neurite formation and was found to be up-regulated after co-cultivation. The differences of Nestin and NF were marginal between co-culture and control cells, but detectable. This could be due to the short incubation time of 48 h which did not allow the cells to completely differentiate into a neuronal cell type. In addition, the expression of αSMA, which indicates inhibition of mesodermal differentiation and, therefore, a reduction provides further evidence for a guided differentiation into the neuronal lineage, was not affected by co-culture.
During co-culture experiments, nasal polyp cells and rat brain biopsies shared the same cultivation media, but were separated through a porous membrane, so that only soluble factors unleashed from the rat brain could affect the stem cell differentiation. Especially factors from necrotic nerve tissue sections can induce neuronal differentiation in adult stem cells (Anwar et al. 2008). Due to this, we analyzed the media supernatants and detected bFGF in the rat brain only approaches, which has neurogenic properties (Komobuchi et al. 2010). The nasal polyp cells themselves expressed IGFBP-2 and IGFBP-6 and were related to adult stem cells, which underlined the existence of stem cells in nasal polyps (Huynh et al. 2011; Petschnik et al. 2011; Ramalho-Santos et al. 2002). Due to the use of FBS, factors which were already present in the culture media were excluded from the analysis. When we excluded all these factors from the rat brain biopsies, nasal polyp cells, and culture media, there is only one factor we found in the supernatants of the co-culture experiments, namely GM-CSF. Under normal conditions, GM-CSF is a cytokine that induces the proliferation, chemotactic migration of astrocytes, and microglia, and has a neuroprotective effect (Guillemin et al. 1996; Lee et al. 1994; Schäbitz et al. 2008), but it was also secreted by human oligodendrocytes, which were generated from human neural stem cells (Kim et al. 2014). The nasal epithelium is the first barrier against environmental factors like allergens, bacteria, or viruses, and responds by producing cytokines which mediate the recruitment of immunocompetent cells (Vroling et al. 2008). This leads to (chronic) inflammatory conditions and the epithelium must regenerate to restore its defensive functions (Watelet et al. 2006). This regenerative process may involve stem cells with self-renewal and proliferation capacities (Alison and Islam 2009). However, in the current literature, only little is known about the existence of stem cells in nasal polyps. Until now, only a few researchers have addressed this issue. Cho et al. (2015) isolated and characterized MSC in nasal polyps and proposed nasal polyp-derived MSCs as an alternative source of MSCs. Otherwise, Pezato et al. (2014) demonstrated that bone marrow-derived MSCs had an immunoregulatory effect on immune cells (especially T cells) derived from nasal tissue affected by CRSwNP. Furthermore, MSCs appeared to have healing modulation properties, which impaired MSCs in the remodeling process (Pezato et al. 2016a), but further studies have to be done to clarify the importance of MSCs in the pathogenesis of nasal polyps. Besides MSCs, Nestin, and BMI-1, two candidates for stem cell markers and renewal factors in human nasal mucosa were localized to the epithelium and submucosal glands of normal nasal mucosa and nasal polyps (Kim et al. 2009). In addition, basal cells in the nasal epithelium showed stemness/progenitor characters with reduced growth/proliferation dynamics (Yu et al. 2014).
In conclusion, our study demonstrated the expression of stem cell-related markers in nasal polyps. Furthermore, we characterized, for the first time, the stemness and neuronal differentiation potential of nasal polyp cells. However, further studies are necessary to validate the function of stem cells in the pathogenesis of nasal polyps in chronic rhinosinusitis, and if so, whether its therapeutic effectiveness could represent a promising strategy in the future.
References
Alison MR, Islam S (2009) Attributes of adult stem cells. J Pathol 217:144–160
Anwar MR, Andreasen CM, Lippert SK et al (2008) Dopaminergic differentiation of human neural stem cells mediated by co-cultured rat striatal brain slices. J Neurochem 105:460–470
Avilion AA, Nicolis SK, Pevny LH et al (2003) Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17:126–140
Ayers MM, Jeffery PK (1988) Proliferation and differentiation in mammalian airway epithelium. Eur Respir J 1:58–80
Campbell PA, Perez-Iratxeta C, Andrade-Navarro MA et al (2007) Oct4 targets regulatory nodes to modulate stem cell function. PLoS One 2:e553
Carden MJ, Trojanowski JQ, Schlaepfer WW et al (1987) Two-stage expression of neurofilament polypeptides during rat neurogenesis with early establishment of adult phosphorylation patterns. J Neurosci 7:3489–3504
Cho KS, Kim YW, Kang MJ et al (2014) Immunomodulatory effect of mesenchymal stem cells on T lymphocyte and cytokine expression in nasal polyps. Otolaryngol Head Neck Surg 150:1062–1070
Cho JS, Park JH, Kang JH et al (2015) Isolation and characterization of multipotent mesenchymal stem cells in nasal polyps. Exp Biol Med 240:185–193
de Borja Callejas F, Martínez-Antón A, Alobid I et al (2014) Reconstituted human upper airway epithelium as 3-d in vitro model for nasal polyposis. PLoS One 9:e100537
Fleige S, Pfaffl MW (2006) RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med 27:126–139
Fokkens WJ, Lund VJ, Mullol J et al (2012) European position paper on rhinosinusitis and nasal polyps 2012. Rhinol Suppl(23):3 (preceding table of contents, 1–298)
Guillemin G, Boussin FD, Le Grand R et al (1996) Granulocyte macrophage colony stimulating factor stimulates in vitro proliferation of astrocytes derived from simian mature brains. Glia 16:71–80
Gupta S, Rosenberg ME (2008) Do stem cells exist in the adult kidney? Am J Nephrol 28:607–613
Huynh H, Zheng J, Umikawa M et al (2011) IGF binding protein 2 supports the survival and cycling of hematopoietic stem cells. Blood 118:3236–3243
Jiang Y, Jahagirdar BN, Reinhardt RL et al (2002) Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 418:41–49
Jordan CT, Guzman ML, Noble M (2006) Cancer stem cells. N Engl J Med 355:1253–1261
Kim YK, Nakagawa N, Nakano K et al (1997) Stem cell factor in nasal polyposis and allergic rhinitis: increased expression by structural cells is suppressed by in vivo topical corticosteroids. J Allergy Clin Immunol 100:389–399
Kim TH, Lee HM, Lee SH et al (2009) Expression and distribution patterns of the stem cell marker, nestin, and the stem cell renewal factor, BMI-1, in normal human nasal mucosa and nasal polyps. Acta Otolaryngol 129:996–1001
Kim WK, Kim D, Cui J et al (2014) Secretome analysis of human oligodendrocytes derived from neural stem cells. PLoS One 9:e84292
Komobuchi H, Hato N, Teraoka M et al (2010) Basic fibroblast growth factor combined with biodegradable hydrogel promotes healing of facial nerve after compression injury: an experimental study. Acta Otolaryngol 130:173–178
Könnecke M, Boscke R, Waldmann A et al (2014) Immune imbalance in nasal polyps of caucasian chronic rhinosinusitis patients is associated with a downregulation of E-selectin. J Immunol Res 2014:959854
Kowalski ML, Lewandowska-Polak A, Woźniak J et al (2005) Association of stem cell factor expression in nasal polyp epithelial cells with aspirin sensitivity and asthma. Allergy 60:631–637
Lee SC, Liu W, Brosnan CF et al (1994) GM-CSF promotes proliferation of human fetal and adult microglia in primary cultures. Glia 12:309–318
Lengner CJ, Welstead GG, Jaenisch R (2008) The pluripotency regulator Oct4: a role in somatic stem cells? Cell Cycle 7:725–728
Li YY, Li CW, Chao SS et al (2014) Impairment of cilia architecture and ciliogenesis in hyperplastic nasal epithelium from nasal polyps. J Allergy Clin Immunol 134:1282–1292
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Nichols J, Zevnik B, Anastassiadis K et al (1998) Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379–391
Pardal R, Clarke MF, Morrison SJ (2003) Applying the principles of stem-cell biology to cancer. Nat Rev Cancer 3:895–902
Petschnik AE, Fell B, Tiede S et al (2011) A novel xenogeneic co-culture system to examine neuronal differentiation capability of various adult human stem cells. PLoS One 6:e24944
Pezato R, de Almeida DC, Bezerra TF et al (2014) Immunoregulatory effects of bone marrow-derived mesenchymal stem cells in the nasal polyp microenvironment. Mediators Inflamm 2014:583409
Pezato R et al (2016a) Hypotheses about the potential role of mesenchymal stem cell on nasal polyposis: a soft inflamed tissue suffering from mechanical dysfunction. Austin Immunol 1(1):1004
Pezato R, Voegels RL, Stamm AC et al (2016b) Why we should avoid using inferior turbinate tissue as control to nasal polyposis studies. Acta Otolaryngol 136:973–975
Ramalho-Santos M, Yoon S, Matsuzaki Y et al (2002) “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 298:597–600
Randell SH (2006) Airway epithelial stem cells and the pathophysiology of chronic obstructive pulmonary disease. Proc Am Thoracic Soc 3:718–725
Reyes M, Dudek A, Jahagirdar B et al (2002) Origin of endothelial progenitors in human postnatal bone marrow. J Clin Invest 109:337–346
Schäbitz WR, Krüger C, Pitzer C et al (2008) A neuroprotective function for the hematopoietic protein granulocyte-macrophage colony stimulating factor (GM-CSF). J Cereb Blood Flow Metab 28:29–43
Vroling AB, Jonker MJ, Luiten S et al (2008) Primary nasal epithelium exposed to house dust mite extract shows activated expression in allergic individuals. Am J Respir Cell Mol Biol 38:293–299
Watelet JB, Van Zele T, Gjomarkaj M et al (2006) Tissue remodelling in upper airways: where is the link with lower airway remodelling? Allergy 61:1249–1258
Wiese C, Rolletschek A, Kania G et al (2004) Nestin expression–a property of multi-lineage progenitor cells? Cell Mol Life Sci 61:2510–2522
Wiszniewski L, Jornot L, Dudez T et al (2006) Long-term cultures of polarized airway epithelial cells from patients with cystic fibrosis. Am J Respir Cell Mol Biol 34:39–48
Yaworsky PJ, Kappen C (1999) Heterogeneity of neural progenitor cells revealed by enhancers in the nestin gene. Dev Biol 205:309–321
Yu H, Fang D, Kumar SM et al (2006) Isolation of a novel population of multipotent adult stem cells from human hair follicles. Am J Pathol 168:1879–1888
Yu F, Zhao X, Li C et al (2012) Airway stem cells: review of potential impact on understanding of upper airway diseases. Laryngoscope 122:1463–1469
Yu XM, Li CW, Chao SS et al (2014) Reduced growth and proliferation dynamics of nasal epithelial stem/progenitor cells in nasal polyps in vitro. Sci Rep 4:4619
Acknowledgements
This work was supported by the Cassella-med GmbH & Co. KG, Köln, Germany and the Rudolf Bartling-Stiftung, Hannover, Germany. We are grateful to all the members of the Department of Otorhinolaryngology for helpful discussions and a comfortable atmosphere as well as Carolin Wienke from the Fraunhofer Research Institution for technical assistance during co-culture experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there were no conflicts of interests regarding the publication of this paper.
Research involving human participants and/or animals
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Informed consent
Informed consent was obtained from all individual participants included in the study.
About this article
Cite this article
Koennecke, M., Böscke, R., Pfannerstill, AC. et al. Neuronal Differentiation Capability of Nasal Polyps of Chronic Rhinosinusitis. Arch. Immunol. Ther. Exp. 65, 431–443 (2017). https://doi.org/10.1007/s00005-017-0456-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00005-017-0456-8