Abstract
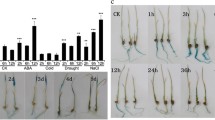
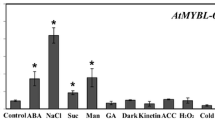
Plants have developed certain adaptive responses to environmental stresses that cause adverse effects on growth. To identify genes involved in the adaptive mechanisms, we constructed a large population of transgenic Arabidopsis expressing rice full-length cDNAs, and performed gain-of-function screening under high-salinity stress. In this study, we identified a rice R2R3-type MYB transcription factor gene, JAmyb, as a gene whose overexpression causes tolerance to high salinity. JAmyb overexpression in transgenic Arabidopsis improved tolerance to high-salinity stress during seed germination, seedling growth, and root elongation. In rice seedlings, JAmyb expression was induced by high-salinity and high-osmotic stresses and reactive oxygen species (ROS), suggesting that JAmyb is responsible for abiotic stress response. Microarray analysis showed that the overexpression of JAmyb stimulates the expression of several defense-associated genes, some of which have been predicted to be involved in osmotic adjustment, ROS removal, and ion homeostasis. Several transcription factors involved in the jasmonate (JA)-mediated stress response are also regulated by JAmyb. JAmyb has been reported to be associated with disease response. Our observations suggest that JAmyb plays a role in JA-mediated abiotic stress response in addition to biotic stress response in rice.



Similar content being viewed by others
Abbreviations
- ABA:
-
Abscisic acid
- JA:
-
Jasmonate
- PQ:
-
Paraquat
- ROS:
-
Reactive oxygen species
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
An Y-Q, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10:107–121
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Baxter CE, Costa MM, Coen ES (2007) Diversification and co-option of RAD-like genes in the evolution of floral asymmetry. Plant J 52:105–113
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300
Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics 19:185–193
Bowler C, Fluhr R (2000) The role of calcium and activated oxygens as signals for controlling cross-tolerance. Trends Plant Sci 5:241–246
Bu Q, Jiang H, Li CB, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C (2008) Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res 18:756–767
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15:1196–1200
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9:436–442
Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K (2011) ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res In press
Hetherington AM, Brownlee C (2004) The generation of Ca2+ signals in plants. Annu Rev Plant Biol 55:401–427
Hundertmark M, Hincha DK (2008) LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9:118e
Ichikawa T, Nakazawa M, Kawashima M, Iizumi H, Kuroda H, Kondou Y, Tsuhara Y, Suzuki K, Ishikawa A, Seki M, Fujita M, Motohashi R, Nagata N, Takagi T, Shinozaki K, Matsui M (2006) The FOX hunting system: an alternative gain-of-function gene hunting technique. Plant J 48:974–985
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264
Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ (2008) Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol 146:623–635
Kondou Y, Higuchi M, Takahashi S, Sakurai T, Ichikawa T et al (2009) Systematic approaches to using the FOX hunting system to identify useful rice genes. Plant J 57:883–894
Kramell R, Atzorn R, Schneider G, Miersch O, Brückner C, Schmidt J, Sembdner G, Parthier B (1995) Occurrence and identification of jasmonic acid and its amino acid conjugates induced by osmotic stress in barley leaf tissue. J Plant Growth Regul 14:29–36
Lee MW, Yang Y (2003) Overexpression of a pathogen-inducible Myb gene increases disease resistance in transgenic rice. B.R. Wells Rice Research Studies. AAES research series, pp 65–72
Lee MW, Qi M, Yang Y (2001) A novel jasmonic acid-inducible rice myb gene associates with fungal infection and host cell death. Mol Plant Microbe Interact 14:527–535
Liang YK, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM (2005) AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol 15:1201–1206
Mandaokar A, Thines B, Shin B, Lange BM, Choi G, Koo YJ, Yoo YJ, Choi YD, Choi G, Browse J (2006) Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J 46:984–1008
McElroy D, Rothenberg M, Reece KS, Wu R (1990) Characterization of the rice (Oryza sativa) actin gene family. Plant Mol Biol 15:257–268
Mengiste T, Chen X, Salmeron J, Dietrich R (2003) The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15:2551–2565
Murgia I, Tarantino D, Soave C, Morandini P (2011) Arabidopsis CYP82C4 expression is dependent on Fe availability and circadian rhythm, and correlates with genes involved in the early Fe deficiency response. J Plant Physiol 168:894–902
Passarinho PA, Van Hengel AJ, Fransz PF, de Vries SC (2001) Expression pattern of the Arabidopsis thaliana AtEP3/AtchitIV endochitinase gene. Planta 212:556–567
Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397:694–697
Rodriguez MC, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61:621–649
Sakurai T, Kondou Y, Akiyama K, Kurotani A, Higuchi M, Ichikawa T, Kuroda H, Kusano M, Mori M, Saitou T, Sakakibara H, Sugano S, Suzuki M, Takahashi H, Takahashi S, Takatsuji H, Yokotani N, Yoshizumi T, Saito K, Shinozaki K, Oda K, Hirochika H, Matsui M (2011) RiceFOX: a database of Arabidopsis mutant lines overexpressing rice full-length cDNA that contains a wide range of trait information to facilitate analysis of gene function. Plant Cell Physiol 52:265–273
Sels J, Mathys J, De Coninck BM, Cammue BP, De Bolle MF (2008) Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol Biochem 46:941–950
Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM (2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23:1138–1115
Singh K, Foley RC, Oñate-Sánchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5:430–436
Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D (2011) The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23:1000–1013
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4:447–456
Tehseen M, Cairns N, Sherson S, Cobbett CS (2010) Metallochaperone-like genes in Arabidopsis thaliana. Metallomics 2:556–564
Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498
Tsonev TD, Lazova GN, Stoinova ZG, Popova LP (1998) A possible role for jasmonic acid in adaptation of barley seedlings to salinity stress. J Plant Growth Regul 17:153–159
Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylidès C, Roby D (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA 99:10179–10184
Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14:1223–1233
Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16:123–132
Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Close TJ (2007) Large-scale expression profiling and physiological characterization of jasmonic acid-mediated adaptation of barley to salinity stress. Plant Cell Environ 30:410–421
Welinder KG, Justesen AF, Kjaersgård IV, Jensen RB, Rasmussen SK, Jespersen HM, Duroux L (2002) Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur J Biochem 269:6063–6081
Wettenhall JM, Simpson KM, Satterley K, Smyth GK (2006) affylmGUI: a graphical user interface for linear modelling of single channel microarray data. Bioinformatics 22:897–899
Xie Z, Li D, Wang L, Sack FD, Grotewold E (2010) Role of the stomatal development regulators FLP/MYB88 in abiotic stress responses. Plant J 64:731–739
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Yokotani N, Ichikawa T, Kondou Y, Maeda S, Iwabuchi M, Mori M, Hirochika H, Matsui M, Oda K (2009) Overexpression of a rice gene encoding a small C2 domain protein OsSMCP1 increases tolerance to abiotic and biotic stresses in transgenic Arabidopsis. Plant Mol Biol 71:391–402
Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18:385–397
Acknowledgments
We thank Dr M. Mori (National Institute of Agrobiological Sciences) for providing the rice seeds. This work was supported in part by Special Coordination Funds for Promoting Science and Technology (Science and Technology Agency of Japan).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yokotani, N., Ichikawa, T., Kondou, Y. et al. Role of the rice transcription factor JAmyb in abiotic stress response. J Plant Res 126, 131–139 (2013). https://doi.org/10.1007/s10265-012-0501-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-012-0501-y