Summary
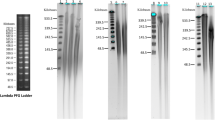
Genomic DNA was isolated fromin vitro Pinus radiata seedling with five DNA isolation protocols commonly used for pines. The methods described by Jobes et al. (1995) and Nelson et al. (1994) utilize sodium dodecyl sulfate, whereas those of Murray and Thompson (1980), Doyle and Doyle (1990), and Devey et al. (1996) use cetyltrimethyl ammonium bromide for cell lysis. The quality and quantity of the isolated DNA was measured and compared. Lithium chloride was found to be more effective than RNase for minimizing the amount of RNA present in the solution. Protocols described by Jobes et al. (1995) and Devey et al. (1996) yielded a large quantity of pure DNA which was suitable for restriction enzyme digestion and polymerase chain reaction amplification. With these methods, 37 to 79 μg of DNA with an A260/280 ratio between 1.7 and 1.9 was obtained from 1 g ofPinus radiata seedlings grownin vitro.
Similar content being viewed by others
References
Ausubel, F. M.; Brent, R.; Kingston, R., et al. Current protocols in molecular biology. New York: John Wiley and Sons; 1987:12–19.
Bousquet, J.; Simon, L.; Lalonde, M. DNA amplification from vegetative and sexual tissues of trees using polymerase chain reaction. Can. J. For. Res. 20:254–257; 1990.
Devey, M. E.; Bell, J. C.; Smith, D. N., et al. A genetic linkage map forPinus radiata based on RFLP, RAPD, and microsatellite markers. Theor. Appl. Genet. 92:673–679; 1996.
Doyle, J. J.; Doyle, J. L. Isolation of plant DNA from fresh tissue. Focus 12:12–15; 1990.
Guries, R. P.; Friedman, S. T.; Ledig, F. T. A megagametophyte analysis of genetic linkage in pitch pine (Pinus rigida Mill.). Heredity 40:309–314; 1978.
Jobes, D. V.; Hurley, D. L.; Thien, L. B. Plant DNA isolation: method to efficiently remove polyphenolics, polysaccharides and RNA. Taxon 44:379–389; 1995.
Keller, G. H.; Manak, M. M. DNA probes. Macmillan Publishers, New York, NY; 1993:70–150.
Muralitharan, M. S.; Stuart, S.; Graham, M. Methods in plant molecular biology techniques. Proceedings of Molecular Biology Workshop, Launceston, Tasmania. Published by Applied Biology, University of Tasmania, Launceston; 1994.
Murray, M. G.; Thompson, W. F. Rapid isolation of high molecular weight DNA. Nucleic Acids Res. 8:4221–4235; 1980.
Nelson, C. D.; Kubisiak, T. L.; Stine, M., et al. A genetic linkage map of longleaf pinePinus palustris Mill.) based on random amplified polymorphic DNAs. J. Hered. 85:433–439; 1994.
Von Arnold, S.; Eriksson, T. A revised medium for growth of pea mesophyll protoplasts. Physiol. Plant. 39:257–260; 1977.
Wagner, D. B.; Furnier, G. R.; Saghai-Maroof, M. A., et al. Chloroplast DNA polymorphisms in lodgepole and jack pines and their hybrids. Proc. Natl. Acad. Sci. USA 84:2097–2100; 1987.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ostrowska, E., Muralitharan, M., Chandler, S. et al. Optimizing conditions for DNA isolation fromPinus radiata . In Vitro Cell.Dev.Biol.-Plant 34, 108–111 (1998). https://doi.org/10.1007/BF02822773
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02822773